Model info
| Transcription factor | Rara | ||||||||
| Model | RARA_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 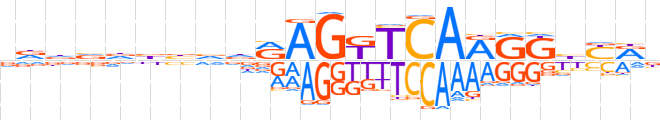 | ||||||||
| LOGO (reverse complement) | 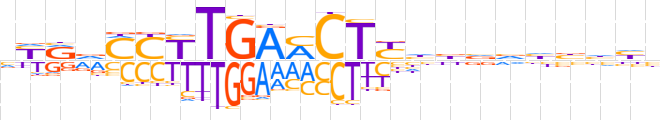 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 23 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | vddvdbbvdRAGKTCARRKbYRb | ||||||||
| Best auROC (human) | 0.863 | ||||||||
| Best auROC (mouse) | 0.945 | ||||||||
| Peak sets in benchmark (human) | 29 | ||||||||
| Peak sets in benchmark (mouse) | 32 | ||||||||
| Aligned words | 498 | ||||||||
| TF family | Thyroid hormone receptor-related factors (NR1) {2.1.2} | ||||||||
| TF subfamily | Retinoic acid receptors (NR1B) {2.1.2.1} | ||||||||
| MGI | MGI:97856 | ||||||||
| EntrezGene | GeneID:19401 (SSTAR profile) | ||||||||
| UniProt ID | RARA_MOUSE | ||||||||
| UniProt AC | P11416 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Rara expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 53.0 | 9.0 | 56.0 | 11.0 | 27.0 | 9.0 | 12.0 | 19.0 | 110.0 | 8.0 | 97.0 | 25.0 | 17.0 | 4.0 | 33.0 | 8.0 |
| 02 | 53.0 | 13.0 | 125.0 | 16.0 | 9.0 | 5.0 | 5.0 | 11.0 | 37.0 | 27.0 | 71.0 | 63.0 | 22.0 | 6.0 | 13.0 | 22.0 |
| 03 | 20.0 | 10.0 | 82.0 | 9.0 | 9.0 | 19.0 | 8.0 | 15.0 | 46.0 | 16.0 | 134.0 | 18.0 | 13.0 | 40.0 | 37.0 | 22.0 |
| 04 | 23.0 | 6.0 | 38.0 | 21.0 | 60.0 | 6.0 | 3.0 | 16.0 | 90.0 | 14.0 | 123.0 | 34.0 | 16.0 | 12.0 | 21.0 | 15.0 |
| 05 | 26.0 | 35.0 | 45.0 | 83.0 | 10.0 | 10.0 | 2.0 | 16.0 | 22.0 | 35.0 | 30.0 | 98.0 | 2.0 | 24.0 | 19.0 | 41.0 |
| 06 | 13.0 | 17.0 | 18.0 | 12.0 | 27.0 | 40.0 | 12.0 | 25.0 | 23.0 | 31.0 | 24.0 | 18.0 | 8.0 | 164.0 | 33.0 | 33.0 |
| 07 | 17.0 | 14.0 | 33.0 | 7.0 | 175.0 | 37.0 | 14.0 | 26.0 | 22.0 | 18.0 | 39.0 | 8.0 | 21.0 | 33.0 | 17.0 | 17.0 |
| 08 | 67.0 | 15.0 | 137.0 | 16.0 | 50.0 | 8.0 | 4.0 | 40.0 | 36.0 | 17.0 | 24.0 | 26.0 | 14.0 | 1.0 | 23.0 | 20.0 |
| 09 | 67.0 | 11.0 | 87.0 | 2.0 | 28.0 | 6.0 | 7.0 | 0.0 | 72.0 | 8.0 | 102.0 | 6.0 | 20.0 | 9.0 | 71.0 | 2.0 |
| 10 | 174.0 | 0.0 | 11.0 | 2.0 | 27.0 | 3.0 | 4.0 | 0.0 | 231.0 | 2.0 | 31.0 | 3.0 | 7.0 | 3.0 | 0.0 | 0.0 |
| 11 | 16.0 | 4.0 | 417.0 | 2.0 | 2.0 | 1.0 | 5.0 | 0.0 | 5.0 | 0.0 | 41.0 | 0.0 | 0.0 | 1.0 | 4.0 | 0.0 |
| 12 | 1.0 | 0.0 | 10.0 | 12.0 | 1.0 | 0.0 | 4.0 | 1.0 | 3.0 | 11.0 | 193.0 | 260.0 | 0.0 | 0.0 | 1.0 | 1.0 |
| 13 | 1.0 | 0.0 | 1.0 | 3.0 | 3.0 | 2.0 | 2.0 | 4.0 | 12.0 | 21.0 | 17.0 | 158.0 | 0.0 | 11.0 | 0.0 | 263.0 |
| 14 | 3.0 | 9.0 | 4.0 | 0.0 | 5.0 | 24.0 | 4.0 | 1.0 | 1.0 | 17.0 | 2.0 | 0.0 | 3.0 | 403.0 | 19.0 | 3.0 |
| 15 | 7.0 | 3.0 | 2.0 | 0.0 | 445.0 | 2.0 | 3.0 | 3.0 | 29.0 | 0.0 | 0.0 | 0.0 | 4.0 | 0.0 | 0.0 | 0.0 |
| 16 | 361.0 | 23.0 | 78.0 | 23.0 | 5.0 | 0.0 | 0.0 | 0.0 | 3.0 | 0.0 | 2.0 | 0.0 | 1.0 | 0.0 | 2.0 | 0.0 |
| 17 | 28.0 | 9.0 | 328.0 | 5.0 | 6.0 | 6.0 | 7.0 | 4.0 | 23.0 | 14.0 | 39.0 | 6.0 | 4.0 | 3.0 | 14.0 | 2.0 |
| 18 | 6.0 | 10.0 | 36.0 | 9.0 | 8.0 | 5.0 | 8.0 | 11.0 | 22.0 | 9.0 | 338.0 | 19.0 | 0.0 | 4.0 | 7.0 | 6.0 |
| 19 | 9.0 | 9.0 | 10.0 | 8.0 | 9.0 | 7.0 | 4.0 | 8.0 | 38.0 | 92.0 | 44.0 | 215.0 | 8.0 | 12.0 | 11.0 | 14.0 |
| 20 | 16.0 | 25.0 | 18.0 | 5.0 | 3.0 | 84.0 | 10.0 | 23.0 | 15.0 | 23.0 | 18.0 | 13.0 | 7.0 | 201.0 | 7.0 | 30.0 |
| 21 | 10.0 | 7.0 | 19.0 | 5.0 | 262.0 | 26.0 | 15.0 | 30.0 | 26.0 | 7.0 | 16.0 | 4.0 | 25.0 | 6.0 | 30.0 | 10.0 |
| 22 | 32.0 | 48.0 | 137.0 | 106.0 | 20.0 | 5.0 | 5.0 | 16.0 | 9.0 | 23.0 | 32.0 | 16.0 | 10.0 | 13.0 | 17.0 | 9.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.527 | -1.211 | 0.582 | -1.018 | -0.14 | -1.211 | -0.934 | -0.486 | 1.254 | -1.324 | 1.128 | -0.216 | -0.595 | -1.971 | 0.058 | -1.324 |
| 02 | 0.527 | -0.856 | 1.381 | -0.654 | -1.211 | -1.766 | -1.766 | -1.018 | 0.171 | -0.14 | 0.818 | 0.699 | -0.342 | -1.596 | -0.856 | -0.342 |
| 03 | -0.435 | -1.11 | 0.961 | -1.211 | -1.211 | -0.486 | -1.324 | -0.717 | 0.387 | -0.654 | 1.45 | -0.539 | -0.856 | 0.248 | 0.171 | -0.342 |
| 04 | -0.298 | -1.596 | 0.197 | -0.388 | 0.65 | -1.596 | -2.23 | -0.654 | 1.054 | -0.784 | 1.365 | 0.087 | -0.654 | -0.934 | -0.388 | -0.717 |
| 05 | -0.177 | 0.116 | 0.365 | 0.973 | -1.11 | -1.11 | -2.58 | -0.654 | -0.342 | 0.116 | -0.036 | 1.139 | -2.58 | -0.256 | -0.486 | 0.273 |
| 06 | -0.856 | -0.595 | -0.539 | -0.934 | -0.14 | 0.248 | -0.934 | -0.216 | -0.298 | -0.004 | -0.256 | -0.539 | -1.324 | 1.652 | 0.058 | 0.058 |
| 07 | -0.595 | -0.784 | 0.058 | -1.451 | 1.717 | 0.171 | -0.784 | -0.177 | -0.342 | -0.539 | 0.223 | -1.324 | -0.388 | 0.058 | -0.595 | -0.595 |
| 08 | 0.76 | -0.717 | 1.472 | -0.654 | 0.469 | -1.324 | -1.971 | 0.248 | 0.144 | -0.595 | -0.256 | -0.177 | -0.784 | -3.122 | -0.298 | -0.435 |
| 09 | 0.76 | -1.018 | 1.02 | -2.58 | -0.104 | -1.596 | -1.451 | -4.397 | 0.832 | -1.324 | 1.178 | -1.596 | -0.435 | -1.211 | 0.818 | -2.58 |
| 10 | 1.711 | -4.397 | -1.018 | -2.58 | -0.14 | -2.23 | -1.971 | -4.397 | 1.994 | -2.58 | -0.004 | -2.23 | -1.451 | -2.23 | -4.397 | -4.397 |
| 11 | -0.654 | -1.971 | 2.584 | -2.58 | -2.58 | -3.122 | -1.766 | -4.397 | -1.766 | -4.397 | 0.273 | -4.397 | -4.397 | -3.122 | -1.971 | -4.397 |
| 12 | -3.122 | -4.397 | -1.11 | -0.934 | -3.122 | -4.397 | -1.971 | -3.122 | -2.23 | -1.018 | 1.814 | 2.112 | -4.397 | -4.397 | -3.122 | -3.122 |
| 13 | -3.122 | -4.397 | -3.122 | -2.23 | -2.23 | -2.58 | -2.58 | -1.971 | -0.934 | -0.388 | -0.595 | 1.615 | -4.397 | -1.018 | -4.397 | 2.123 |
| 14 | -2.23 | -1.211 | -1.971 | -4.397 | -1.766 | -0.256 | -1.971 | -3.122 | -3.122 | -0.595 | -2.58 | -4.397 | -2.23 | 2.549 | -0.486 | -2.23 |
| 15 | -1.451 | -2.23 | -2.58 | -4.397 | 2.649 | -2.58 | -2.23 | -2.23 | -0.07 | -4.397 | -4.397 | -4.397 | -1.971 | -4.397 | -4.397 | -4.397 |
| 16 | 2.44 | -0.298 | 0.911 | -0.298 | -1.766 | -4.397 | -4.397 | -4.397 | -2.23 | -4.397 | -2.58 | -4.397 | -3.122 | -4.397 | -2.58 | -4.397 |
| 17 | -0.104 | -1.211 | 2.344 | -1.766 | -1.596 | -1.596 | -1.451 | -1.971 | -0.298 | -0.784 | 0.223 | -1.596 | -1.971 | -2.23 | -0.784 | -2.58 |
| 18 | -1.596 | -1.11 | 0.144 | -1.211 | -1.324 | -1.766 | -1.324 | -1.018 | -0.342 | -1.211 | 2.374 | -0.486 | -4.397 | -1.971 | -1.451 | -1.596 |
| 19 | -1.211 | -1.211 | -1.11 | -1.324 | -1.211 | -1.451 | -1.971 | -1.324 | 0.197 | 1.076 | 0.343 | 1.922 | -1.324 | -0.934 | -1.018 | -0.784 |
| 20 | -0.654 | -0.216 | -0.539 | -1.766 | -2.23 | 0.985 | -1.11 | -0.298 | -0.717 | -0.298 | -0.539 | -0.856 | -1.451 | 1.855 | -1.451 | -0.036 |
| 21 | -1.11 | -1.451 | -0.486 | -1.766 | 2.119 | -0.177 | -0.717 | -0.036 | -0.177 | -1.451 | -0.654 | -1.971 | -0.216 | -1.596 | -0.036 | -1.11 |
| 22 | 0.027 | 0.429 | 1.472 | 1.217 | -0.435 | -1.766 | -1.766 | -0.654 | -1.211 | -0.298 | 0.027 | -0.654 | -1.11 | -0.856 | -0.595 | -1.211 |