Model info
| Transcription factor | RARG (GeneCards) | ||||||||
| Model | RARG_HUMAN.H11MO.2.D | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO | 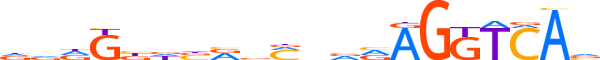 | ||||||||
| LOGO (reverse complement) | 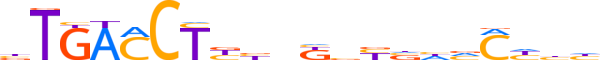 | ||||||||
| Data source | Integrative | ||||||||
| Model release | HOCOMOCOv9 | ||||||||
| Model length | 20 | ||||||||
| Quality | D | ||||||||
| Motif rank | 2 | ||||||||
| Consensus | dddGbdbRvMnvRAGGTCAv | ||||||||
| Best auROC (human) | 0.656 | ||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | 12 | ||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 24 | ||||||||
| TF family | Thyroid hormone receptor-related factors (NR1) {2.1.2} | ||||||||
| TF subfamily | Retinoic acid receptors (NR1B) {2.1.2.1} | ||||||||
| HGNC | HGNC:9866 | ||||||||
| EntrezGene | GeneID:5916 (SSTAR profile) | ||||||||
| UniProt ID | RARG_HUMAN | ||||||||
| UniProt AC | P13631 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | RARG expression | ||||||||
| ReMap ChIP-seq dataset list | RARG datasets | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 4.957 | 2.478 | 12.392 | 4.203 |
| 02 | 5.065 | 3.664 | 11.315 | 3.987 |
| 03 | 7.866 | 0.97 | 11.315 | 3.879 |
| 04 | 0.0 | 0.97 | 18.534 | 4.526 |
| 05 | 1.078 | 4.634 | 10.453 | 7.866 |
| 06 | 5.496 | 1.94 | 4.957 | 11.638 |
| 07 | 2.694 | 13.039 | 4.741 | 3.556 |
| 08 | 12.716 | 3.017 | 7.22 | 1.078 |
| 09 | 5.28 | 8.082 | 7.866 | 2.802 |
| 10 | 4.095 | 15.302 | 2.694 | 1.94 |
| 11 | 5.603 | 7.543 | 7.004 | 3.879 |
| 12 | 11.853 | 3.125 | 6.466 | 2.586 |
| 13 | 8.728 | 5.927 | 9.375 | 0.0 |
| 14 | 20.151 | 0.0 | 3.017 | 0.862 |
| 15 | 0.0 | 0.0 | 24.03 | 0.0 |
| 16 | 0.0 | 0.0 | 16.918 | 7.112 |
| 17 | 4.418 | 0.0 | 0.0 | 19.612 |
| 18 | 2.047 | 20.366 | 1.616 | 0.0 |
| 19 | 22.953 | 1.078 | 0.0 | 0.0 |
| 20 | 6.816 | 8.217 | 7.786 | 1.212 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.168 | -0.731 | 0.662 | -0.308 |
| 02 | -0.149 | -0.422 | 0.577 | -0.352 |
| 03 | 0.242 | -1.349 | 0.577 | -0.375 |
| 04 | -2.147 | -1.349 | 1.044 | -0.246 |
| 05 | -1.29 | -0.226 | 0.503 | 0.242 |
| 06 | -0.078 | -0.911 | -0.168 | 0.603 |
| 07 | -0.668 | 0.71 | -0.206 | -0.447 |
| 08 | 0.686 | -0.579 | 0.164 | -1.29 |
| 09 | -0.113 | 0.266 | 0.242 | -0.637 |
| 10 | -0.33 | 0.861 | -0.668 | -0.911 |
| 11 | -0.061 | 0.204 | 0.137 | -0.375 |
| 12 | 0.62 | -0.551 | 0.065 | -0.699 |
| 13 | 0.336 | -0.012 | 0.402 | -2.147 |
| 14 | 1.125 | -2.147 | -0.579 | -1.412 |
| 15 | -2.147 | -2.147 | 1.295 | -2.147 |
| 16 | -2.147 | -2.147 | 0.957 | 0.15 |
| 17 | -0.266 | -2.147 | -2.147 | 1.099 |
| 18 | -0.873 | 1.135 | -1.037 | -2.147 |
| 19 | 1.25 | -1.29 | -2.147 | -2.147 |
| 20 | 0.112 | 0.281 | 0.232 | -1.221 |