Model info
| Transcription factor | Rfx1 | ||||||||
| Model | RFX1_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 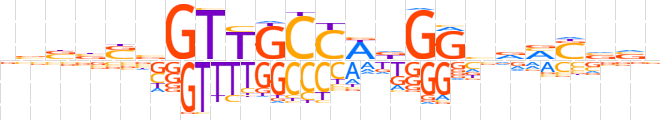 | ||||||||
| LOGO (reverse complement) | 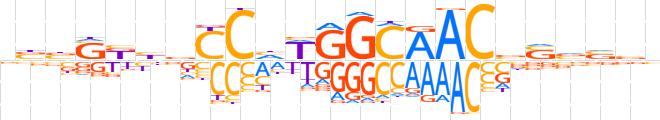 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv10 | ||||||||
| Model length | 23 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | bbbbSbGTTGCCWdGGvvMSvvb | ||||||||
| Best auROC (human) | 0.699 | ||||||||
| Best auROC (mouse) | 0.994 | ||||||||
| Peak sets in benchmark (human) | 3 | ||||||||
| Peak sets in benchmark (mouse) | 7 | ||||||||
| Aligned words | 440 | ||||||||
| TF family | RFX-related factors {3.3.3} | ||||||||
| TF subfamily | RFX1 (EF-C) {3.3.3.0.1} | ||||||||
| MGI | MGI:105982 | ||||||||
| EntrezGene | GeneID:19724 (SSTAR profile) | ||||||||
| UniProt ID | RFX1_MOUSE | ||||||||
| UniProt AC | P48377 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Rfx1 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 7.143 | 12.594 | 21.655 | 3.324 | 17.32 | 56.396 | 49.266 | 39.135 | 25.507 | 100.143 | 35.209 | 16.613 | 5.022 | 15.196 | 23.722 | 12.37 |
| 02 | 1.826 | 20.44 | 29.839 | 2.886 | 9.335 | 91.592 | 47.386 | 36.016 | 8.758 | 61.49 | 43.353 | 16.25 | 3.997 | 32.414 | 24.562 | 10.468 |
| 03 | 1.916 | 8.377 | 11.588 | 2.034 | 18.056 | 74.152 | 68.902 | 44.826 | 11.974 | 55.578 | 60.506 | 17.083 | 2.767 | 23.543 | 29.599 | 9.711 |
| 04 | 1.84 | 19.635 | 9.104 | 4.135 | 9.314 | 88.885 | 45.153 | 18.298 | 6.404 | 112.155 | 42.022 | 10.013 | 1.888 | 37.117 | 21.016 | 13.633 |
| 05 | 1.043 | 7.929 | 10.473 | 0.0 | 42.893 | 74.157 | 86.12 | 54.623 | 6.969 | 61.201 | 39.26 | 9.866 | 4.845 | 7.059 | 30.214 | 3.961 |
| 06 | 1.026 | 1.04 | 53.684 | 0.0 | 4.104 | 0.0 | 146.242 | 0.0 | 0.0 | 0.0 | 166.067 | 0.0 | 0.98 | 0.0 | 66.472 | 0.999 |
| 07 | 0.0 | 0.0 | 0.0 | 6.109 | 0.0 | 0.0 | 0.0 | 1.04 | 1.956 | 16.4 | 2.044 | 412.065 | 0.0 | 0.0 | 0.0 | 0.999 |
| 08 | 0.0 | 1.046 | 0.0 | 0.909 | 1.061 | 5.643 | 0.0 | 9.695 | 1.016 | 1.028 | 0.0 | 0.0 | 5.917 | 74.804 | 1.885 | 337.606 |
| 09 | 3.1 | 0.0 | 4.895 | 0.0 | 1.895 | 1.044 | 64.769 | 14.814 | 0.0 | 0.0 | 0.926 | 0.959 | 13.817 | 4.223 | 310.773 | 19.399 |
| 10 | 0.0 | 18.811 | 0.0 | 0.0 | 0.0 | 5.267 | 0.0 | 0.0 | 0.0 | 345.161 | 2.871 | 33.331 | 0.0 | 35.172 | 0.0 | 0.0 |
| 11 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 316.195 | 1.768 | 86.448 | 1.058 | 1.813 | 0.0 | 0.0 | 0.0 | 29.351 | 0.0 | 3.98 |
| 12 | 0.0 | 0.0 | 1.058 | 0.0 | 248.188 | 26.894 | 25.527 | 46.75 | 0.901 | 0.0 | 0.867 | 0.0 | 75.258 | 1.791 | 12.224 | 1.155 |
| 13 | 45.391 | 15.209 | 89.111 | 174.636 | 2.717 | 4.311 | 10.834 | 10.823 | 3.74 | 4.997 | 19.039 | 11.9 | 15.936 | 2.17 | 16.473 | 13.325 |
| 14 | 1.964 | 0.0 | 65.82 | 0.0 | 3.955 | 0.0 | 22.732 | 0.0 | 2.968 | 0.872 | 131.618 | 0.0 | 12.277 | 0.0 | 198.406 | 0.0 |
| 15 | 0.0 | 0.0 | 21.164 | 0.0 | 0.0 | 0.0 | 0.872 | 0.0 | 50.222 | 21.939 | 340.565 | 5.851 | 0.0 | 0.0 | 0.0 | 0.0 |
| 16 | 0.935 | 25.059 | 22.197 | 2.031 | 12.114 | 1.98 | 5.887 | 1.957 | 68.42 | 162.39 | 68.414 | 63.376 | 1.942 | 0.862 | 3.047 | 0.0 |
| 17 | 27.773 | 3.762 | 50.826 | 1.051 | 79.724 | 15.509 | 74.967 | 20.092 | 58.022 | 22.077 | 15.364 | 4.082 | 20.429 | 4.66 | 26.497 | 15.778 |
| 18 | 87.184 | 48.409 | 44.798 | 5.557 | 19.888 | 5.797 | 17.453 | 2.87 | 131.068 | 25.243 | 9.441 | 1.902 | 22.017 | 5.061 | 6.866 | 7.059 |
| 19 | 15.576 | 221.376 | 8.007 | 15.198 | 9.071 | 40.125 | 22.758 | 12.556 | 5.254 | 51.59 | 14.893 | 6.82 | 4.066 | 9.371 | 2.021 | 1.93 |
| 20 | 4.939 | 18.097 | 9.097 | 1.834 | 63.802 | 97.085 | 136.329 | 25.246 | 3.834 | 10.861 | 30.702 | 2.282 | 2.265 | 1.93 | 28.579 | 3.73 |
| 21 | 18.072 | 12.094 | 41.36 | 3.313 | 28.998 | 26.022 | 59.32 | 13.633 | 26.618 | 69.719 | 100.64 | 7.731 | 4.331 | 7.438 | 20.38 | 0.943 |
| 22 | 11.564 | 20.259 | 32.978 | 13.218 | 19.735 | 27.691 | 39.787 | 28.06 | 20.176 | 90.359 | 78.131 | 33.035 | 4.721 | 9.007 | 7.934 | 3.958 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -1.311 | -0.766 | -0.237 | -2.02 | -0.456 | 0.71 | 0.576 | 0.347 | -0.076 | 1.281 | 0.243 | -0.496 | -1.643 | -0.584 | -0.147 | -0.784 |
| 02 | -2.538 | -0.293 | 0.079 | -2.146 | -1.056 | 1.192 | 0.537 | 0.265 | -1.117 | 0.796 | 0.449 | -0.518 | -1.853 | 0.161 | -0.113 | -0.945 |
| 03 | -2.498 | -1.159 | -0.847 | -2.448 | -0.415 | 0.982 | 0.909 | 0.482 | -0.815 | 0.695 | 0.78 | -0.469 | -2.183 | -0.154 | 0.071 | -1.018 |
| 04 | -2.532 | -0.333 | -1.08 | -1.822 | -1.058 | 1.162 | 0.489 | -0.402 | -1.415 | 1.394 | 0.418 | -0.988 | -2.51 | 0.295 | -0.266 | -0.689 |
| 05 | -2.976 | -1.212 | -0.945 | -4.296 | 0.438 | 0.982 | 1.131 | 0.678 | -1.335 | 0.791 | 0.351 | -1.002 | -1.676 | -1.323 | 0.092 | -1.861 |
| 06 | -2.988 | -2.979 | 0.661 | -4.296 | -1.829 | -4.296 | 1.659 | -4.296 | -4.296 | -4.296 | 1.785 | -4.296 | -3.022 | -4.296 | 0.873 | -3.008 |
| 07 | -4.296 | -4.296 | -4.296 | -1.459 | -4.296 | -4.296 | -4.296 | -2.979 | -2.481 | -0.509 | -2.444 | 2.693 | -4.296 | -4.296 | -4.296 | -3.008 |
| 08 | -4.296 | -2.974 | -4.296 | -3.075 | -2.963 | -1.534 | -4.296 | -1.019 | -2.995 | -2.987 | -4.296 | -4.296 | -1.489 | 0.991 | -2.511 | 2.494 |
| 09 | -2.082 | -4.296 | -1.666 | -4.296 | -2.507 | -2.976 | 0.847 | -0.608 | -4.296 | -4.296 | -3.062 | -3.037 | -0.676 | -1.803 | 2.411 | -0.345 |
| 10 | -4.296 | -0.375 | -4.296 | -4.296 | -4.296 | -1.598 | -4.296 | -4.296 | -4.296 | 2.516 | -2.15 | 0.189 | -4.296 | 0.242 | -4.296 | -4.296 |
| 11 | -4.296 | -4.296 | -4.296 | -4.296 | -4.296 | 2.428 | -2.564 | 1.135 | -2.966 | -2.544 | -4.296 | -4.296 | -4.296 | 0.063 | -4.296 | -1.857 |
| 12 | -4.296 | -4.296 | -2.966 | -4.296 | 2.186 | -0.023 | -0.075 | 0.524 | -3.081 | -4.296 | -3.108 | -4.296 | 0.997 | -2.554 | -0.795 | -2.9 |
| 13 | 0.494 | -0.583 | 1.165 | 1.836 | -2.199 | -1.784 | -0.912 | -0.913 | -1.913 | -1.647 | -0.363 | -0.821 | -0.537 | -2.393 | -0.505 | -0.711 |
| 14 | -2.477 | -4.296 | 0.863 | -4.296 | -1.862 | -4.296 | -0.189 | -4.296 | -2.121 | -3.105 | 1.553 | -4.296 | -0.791 | -4.296 | 1.963 | -4.296 |
| 15 | -4.296 | -4.296 | -0.259 | -4.296 | -4.296 | -4.296 | -3.105 | -4.296 | 0.595 | -0.224 | 2.502 | -1.5 | -4.296 | -4.296 | -4.296 | -4.296 |
| 16 | -3.055 | -0.093 | -0.212 | -2.449 | -0.804 | -2.47 | -1.494 | -2.48 | 0.902 | 1.763 | 0.902 | 0.826 | -2.487 | -3.112 | -2.097 | -4.296 |
| 17 | 0.008 | -1.908 | 0.607 | -2.971 | 1.054 | -0.564 | 0.993 | -0.31 | 0.738 | -0.218 | -0.573 | -1.834 | -0.294 | -1.712 | -0.038 | -0.547 |
| 18 | 1.143 | 0.558 | 0.481 | -1.548 | -0.32 | -1.508 | -0.448 | -2.151 | 1.549 | -0.086 | -1.045 | -2.504 | -0.22 | -1.635 | -1.349 | -1.322 |
| 19 | -0.559 | 2.072 | -1.203 | -0.583 | -1.083 | 0.372 | -0.188 | -0.769 | -1.6 | 0.621 | -0.603 | -1.355 | -1.837 | -1.052 | -2.453 | -2.492 |
| 20 | -1.658 | -0.413 | -1.08 | -2.534 | 0.832 | 1.25 | 1.589 | -0.086 | -1.891 | -0.91 | 0.107 | -2.35 | -2.357 | -2.492 | 0.037 | -1.916 |
| 21 | -0.414 | -0.806 | 0.402 | -2.023 | 0.051 | -0.056 | 0.76 | -0.689 | -0.033 | 0.921 | 1.286 | -1.236 | -1.779 | -1.273 | -0.296 | -3.049 |
| 22 | -0.849 | -0.302 | 0.178 | -0.719 | -0.328 | 0.005 | 0.364 | 0.019 | -0.306 | 1.179 | 1.034 | 0.18 | -1.7 | -1.09 | -1.211 | -1.862 |