Model info
| Transcription factor | RFX5 (GeneCards) | ||||||||
| Model | RFX5_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 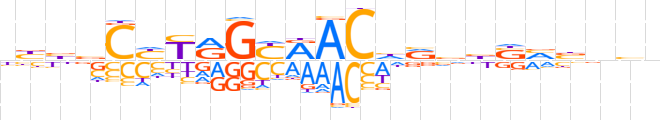 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv10 | ||||||||
| Model length | 23 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nvnvbSvbYdGTYRCYRRGbvvv | ||||||||
| Best auROC (human) | 0.987 | ||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | 14 | ||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 500 | ||||||||
| TF family | RFX-related factors {3.3.3} | ||||||||
| TF subfamily | RFX5 {3.3.3.0.5} | ||||||||
| HGNC | HGNC:9986 | ||||||||
| EntrezGene | GeneID:5993 (SSTAR profile) | ||||||||
| UniProt ID | RFX5_HUMAN | ||||||||
| UniProt AC | P48382 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | RFX5 expression | ||||||||
| ReMap ChIP-seq dataset list | RFX5 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 18.414 | 15.661 | 46.892 | 11.619 | 21.944 | 17.297 | 35.794 | 33.519 | 60.185 | 29.212 | 45.298 | 19.33 | 12.005 | 24.534 | 90.9 | 17.395 |
| 02 | 13.423 | 59.938 | 19.053 | 20.135 | 18.923 | 26.149 | 16.452 | 25.18 | 74.007 | 68.694 | 43.906 | 32.277 | 11.867 | 28.387 | 20.697 | 20.912 |
| 03 | 11.174 | 45.866 | 50.507 | 10.673 | 53.196 | 22.726 | 91.627 | 15.619 | 15.583 | 39.644 | 39.83 | 5.05 | 5.835 | 13.161 | 71.355 | 8.154 |
| 04 | 2.024 | 10.361 | 22.751 | 50.652 | 17.859 | 22.289 | 44.304 | 36.946 | 13.348 | 25.056 | 41.003 | 173.913 | 1.932 | 8.637 | 14.812 | 14.115 |
| 05 | 5.582 | 16.766 | 9.985 | 2.829 | 13.814 | 23.741 | 15.551 | 13.236 | 19.282 | 43.254 | 28.479 | 31.855 | 17.777 | 227.807 | 21.606 | 8.436 |
| 06 | 18.61 | 3.197 | 23.744 | 10.904 | 221.4 | 33.263 | 30.189 | 26.715 | 8.794 | 31.323 | 19.986 | 15.519 | 3.498 | 28.545 | 15.536 | 8.776 |
| 07 | 7.16 | 91.394 | 57.001 | 96.747 | 26.314 | 26.521 | 25.243 | 18.25 | 8.031 | 32.594 | 37.073 | 11.757 | 5.66 | 17.295 | 30.756 | 8.205 |
| 08 | 3.877 | 32.065 | 4.615 | 6.608 | 18.997 | 83.166 | 22.582 | 43.058 | 10.466 | 101.36 | 20.878 | 17.37 | 3.49 | 82.542 | 21.923 | 27.004 |
| 09 | 7.306 | 5.526 | 12.862 | 11.136 | 85.875 | 23.982 | 32.019 | 157.258 | 24.123 | 9.855 | 16.494 | 19.526 | 19.574 | 1.69 | 34.028 | 38.748 |
| 10 | 4.691 | 0.0 | 132.186 | 0.0 | 2.343 | 0.777 | 37.932 | 0.0 | 0.0 | 1.35 | 93.27 | 0.782 | 7.092 | 0.756 | 217.406 | 1.413 |
| 11 | 0.0 | 0.0 | 0.968 | 13.158 | 0.0 | 0.777 | 0.0 | 2.106 | 1.893 | 24.678 | 20.205 | 434.019 | 0.0 | 0.0 | 0.0 | 2.195 |
| 12 | 0.0 | 0.772 | 1.121 | 0.0 | 0.0 | 1.646 | 5.915 | 17.893 | 8.716 | 0.858 | 1.73 | 9.87 | 42.658 | 70.81 | 7.484 | 330.525 |
| 13 | 10.609 | 3.372 | 24.54 | 12.853 | 30.82 | 0.774 | 18.819 | 23.674 | 0.0 | 0.0 | 12.548 | 3.703 | 46.35 | 14.319 | 278.769 | 18.851 |
| 14 | 0.0 | 86.792 | 0.988 | 0.0 | 1.983 | 15.406 | 1.076 | 0.0 | 5.191 | 294.291 | 18.479 | 16.716 | 0.898 | 55.582 | 1.636 | 0.964 |
| 15 | 0.0 | 4.544 | 1.724 | 1.803 | 7.747 | 212.113 | 8.216 | 223.994 | 1.748 | 13.88 | 2.165 | 4.385 | 1.498 | 9.787 | 0.0 | 6.394 |
| 16 | 3.777 | 0.767 | 4.719 | 1.729 | 191.31 | 4.462 | 33.067 | 11.486 | 7.241 | 0.0 | 4.865 | 0.0 | 131.989 | 4.615 | 95.936 | 4.037 |
| 17 | 58.081 | 18.826 | 182.064 | 75.346 | 7.044 | 2.018 | 0.782 | 0.0 | 19.601 | 3.878 | 108.601 | 6.507 | 3.324 | 2.624 | 9.264 | 2.04 |
| 18 | 0.886 | 3.798 | 77.655 | 5.712 | 8.986 | 6.056 | 8.767 | 3.537 | 3.473 | 12.932 | 279.327 | 4.979 | 6.273 | 1.995 | 73.543 | 2.081 |
| 19 | 3.539 | 5.603 | 5.997 | 4.48 | 6.22 | 3.025 | 3.505 | 12.032 | 54.976 | 179.187 | 146.858 | 58.27 | 2.371 | 12.133 | 0.935 | 0.87 |
| 20 | 24.702 | 12.121 | 25.566 | 4.717 | 122.285 | 16.839 | 40.737 | 20.087 | 57.757 | 42.566 | 34.505 | 22.466 | 29.34 | 9.974 | 32.272 | 4.066 |
| 21 | 55.394 | 8.888 | 165.568 | 4.235 | 32.453 | 17.975 | 15.833 | 15.239 | 43.125 | 23.715 | 51.909 | 14.331 | 5.122 | 10.503 | 27.618 | 8.092 |
| 22 | 71.554 | 18.224 | 34.035 | 12.282 | 12.318 | 16.084 | 15.346 | 17.334 | 130.078 | 45.495 | 67.217 | 18.138 | 7.616 | 9.075 | 12.043 | 13.163 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.52 | -0.679 | 0.402 | -0.969 | -0.348 | -0.582 | 0.134 | 0.069 | 0.649 | -0.067 | 0.367 | -0.473 | -0.937 | -0.239 | 1.06 | -0.576 |
| 02 | -0.829 | 0.645 | -0.487 | -0.433 | -0.494 | -0.176 | -0.631 | -0.213 | 0.855 | 0.781 | 0.336 | 0.032 | -0.948 | -0.095 | -0.406 | -0.396 |
| 03 | -1.007 | 0.38 | 0.475 | -1.051 | 0.527 | -0.314 | 1.068 | -0.681 | -0.684 | 0.235 | 0.24 | -1.761 | -1.626 | -0.848 | 0.819 | -1.309 |
| 04 | -2.574 | -1.079 | -0.313 | 0.478 | -0.55 | -0.333 | 0.345 | 0.166 | -0.834 | -0.218 | 0.269 | 1.706 | -2.613 | -1.254 | -0.733 | -0.78 |
| 05 | -1.668 | -0.612 | -1.115 | -2.286 | -0.801 | -0.271 | -0.686 | -0.842 | -0.475 | 0.322 | -0.092 | 0.019 | -0.555 | 1.976 | -0.364 | -1.277 |
| 06 | -0.51 | -2.178 | -0.271 | -1.03 | 1.947 | 0.062 | -0.034 | -0.155 | -1.237 | 0.002 | -0.44 | -0.688 | -2.097 | -0.089 | -0.687 | -1.239 |
| 07 | -1.433 | 1.065 | 0.595 | 1.122 | -0.17 | -0.162 | -0.211 | -0.529 | -1.324 | 0.042 | 0.169 | -0.957 | -1.655 | -0.582 | -0.016 | -1.303 |
| 08 | -2.004 | 0.025 | -1.844 | -1.509 | -0.49 | 0.971 | -0.32 | 0.317 | -1.07 | 1.168 | -0.397 | -0.578 | -2.099 | 0.964 | -0.349 | -0.144 |
| 09 | -1.414 | -1.677 | -0.87 | -1.01 | 1.003 | -0.261 | 0.024 | 1.606 | -0.255 | -1.128 | -0.628 | -0.463 | -0.461 | -2.723 | 0.084 | 0.213 |
| 10 | -1.829 | -4.4 | 1.433 | -4.4 | -2.45 | -3.301 | 0.192 | -4.4 | -4.4 | -2.901 | 1.085 | -3.297 | -1.442 | -3.319 | 1.929 | -2.866 |
| 11 | -4.4 | -4.4 | -3.149 | -0.848 | -4.4 | -3.301 | -4.4 | -2.54 | -2.63 | -0.233 | -0.429 | 2.62 | -4.4 | -4.4 | -4.4 | -2.505 |
| 12 | -4.4 | -3.305 | -3.043 | -4.4 | -4.4 | -2.744 | -1.613 | -0.548 | -1.246 | -3.234 | -2.704 | -1.126 | 0.308 | 0.811 | -1.391 | 2.347 |
| 13 | -1.057 | -2.13 | -0.238 | -0.871 | -0.014 | -3.304 | -0.499 | -0.274 | -4.4 | -4.4 | -0.894 | -2.046 | 0.39 | -0.766 | 2.177 | -0.497 |
| 14 | -4.4 | 1.014 | -3.135 | -4.4 | -2.591 | -0.695 | -3.073 | -4.4 | -1.735 | 2.232 | -0.517 | -0.615 | -3.202 | 0.57 | -2.749 | -3.153 |
| 15 | -4.4 | -1.858 | -2.706 | -2.67 | -1.358 | 1.905 | -1.302 | 1.959 | -2.695 | -0.796 | -2.517 | -1.891 | -2.82 | -1.134 | -4.4 | -1.54 |
| 16 | -2.028 | -3.31 | -1.824 | -2.704 | 1.802 | -1.875 | 0.056 | -0.98 | -1.422 | -4.4 | -1.796 | -4.4 | 1.431 | -1.844 | 1.113 | -1.967 |
| 17 | 0.614 | -0.499 | 1.752 | 0.873 | -1.448 | -2.576 | -3.297 | -4.4 | -0.459 | -2.004 | 1.237 | -1.523 | -2.143 | -2.352 | -1.187 | -2.567 |
| 18 | -3.212 | -2.022 | 0.903 | -1.646 | -1.216 | -1.591 | -1.24 | -2.087 | -2.103 | -0.865 | 2.179 | -1.774 | -1.558 | -2.586 | 0.849 | -2.55 |
| 19 | -2.086 | -1.664 | -1.6 | -1.872 | -1.566 | -2.227 | -2.095 | -0.935 | 0.56 | 1.736 | 1.538 | 0.617 | -2.439 | -0.927 | -3.174 | -3.225 |
| 20 | -0.232 | -0.928 | -0.198 | -1.824 | 1.355 | -0.608 | 0.262 | -0.435 | 0.609 | 0.306 | 0.098 | -0.325 | -0.062 | -1.116 | 0.032 | -1.961 |
| 21 | 0.567 | -1.227 | 1.657 | -1.923 | 0.037 | -0.544 | -0.668 | -0.705 | 0.319 | -0.272 | 0.503 | -0.765 | -1.748 | -1.066 | -0.122 | -1.317 |
| 22 | 0.821 | -0.531 | 0.084 | -0.915 | -0.912 | -0.653 | -0.699 | -0.58 | 1.417 | 0.372 | 0.759 | -0.535 | -1.374 | -1.207 | -0.934 | -0.848 |