Model info
| Transcription factor | Rfx6 | ||||||||
| Model | RFX6_MOUSE.H11DI.0.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 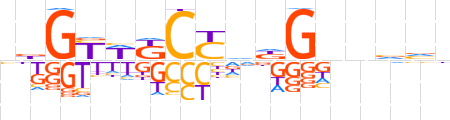 | ||||||||
| LOGO (reverse complement) | 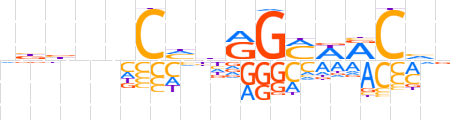 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 16 | ||||||||
| Quality | C | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | bbGYWKCYvdGnnvvn | ||||||||
| Best auROC (human) | |||||||||
| Best auROC (mouse) | 0.954 | ||||||||
| Peak sets in benchmark (human) | |||||||||
| Peak sets in benchmark (mouse) | 3 | ||||||||
| Aligned words | 522 | ||||||||
| TF family | RFX-related factors {3.3.3} | ||||||||
| TF subfamily | RFX6 (RFXDC-1) {3.3.3.0.6} | ||||||||
| MGI | MGI:2445208 | ||||||||
| EntrezGene | GeneID:320995 (SSTAR profile) | ||||||||
| UniProt ID | RFX6_MOUSE | ||||||||
| UniProt AC | Q8C7R7 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Rfx6 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 8.0 | 6.0 | 20.0 | 19.0 | 23.0 | 21.0 | 24.0 | 97.0 | 6.0 | 18.0 | 50.0 | 34.0 | 7.0 | 5.0 | 47.0 | 17.0 |
| 02 | 1.0 | 0.0 | 41.0 | 2.0 | 4.0 | 1.0 | 44.0 | 1.0 | 4.0 | 0.0 | 136.0 | 1.0 | 3.0 | 0.0 | 163.0 | 1.0 |
| 03 | 0.0 | 1.0 | 4.0 | 7.0 | 0.0 | 0.0 | 0.0 | 1.0 | 23.0 | 49.0 | 32.0 | 280.0 | 0.0 | 3.0 | 0.0 | 2.0 |
| 04 | 3.0 | 2.0 | 2.0 | 16.0 | 10.0 | 1.0 | 2.0 | 40.0 | 11.0 | 7.0 | 1.0 | 17.0 | 45.0 | 42.0 | 16.0 | 187.0 |
| 05 | 10.0 | 1.0 | 37.0 | 21.0 | 6.0 | 0.0 | 9.0 | 37.0 | 0.0 | 1.0 | 10.0 | 10.0 | 19.0 | 1.0 | 190.0 | 50.0 |
| 06 | 0.0 | 35.0 | 0.0 | 0.0 | 0.0 | 3.0 | 0.0 | 0.0 | 2.0 | 231.0 | 4.0 | 9.0 | 0.0 | 114.0 | 1.0 | 3.0 |
| 07 | 0.0 | 0.0 | 1.0 | 1.0 | 1.0 | 228.0 | 0.0 | 154.0 | 0.0 | 3.0 | 1.0 | 1.0 | 0.0 | 6.0 | 1.0 | 5.0 |
| 08 | 0.0 | 0.0 | 0.0 | 1.0 | 102.0 | 54.0 | 20.0 | 61.0 | 3.0 | 0.0 | 0.0 | 0.0 | 60.0 | 23.0 | 67.0 | 11.0 |
| 09 | 33.0 | 1.0 | 77.0 | 54.0 | 21.0 | 7.0 | 18.0 | 31.0 | 10.0 | 1.0 | 64.0 | 12.0 | 27.0 | 4.0 | 35.0 | 7.0 |
| 10 | 1.0 | 0.0 | 89.0 | 1.0 | 0.0 | 0.0 | 12.0 | 1.0 | 3.0 | 3.0 | 187.0 | 1.0 | 5.0 | 0.0 | 99.0 | 0.0 |
| 11 | 0.0 | 0.0 | 8.0 | 1.0 | 0.0 | 1.0 | 1.0 | 1.0 | 77.0 | 76.0 | 146.0 | 88.0 | 0.0 | 1.0 | 2.0 | 0.0 |
| 12 | 4.0 | 19.0 | 42.0 | 12.0 | 30.0 | 11.0 | 10.0 | 27.0 | 50.0 | 43.0 | 33.0 | 31.0 | 20.0 | 9.0 | 33.0 | 28.0 |
| 13 | 37.0 | 17.0 | 40.0 | 10.0 | 54.0 | 14.0 | 7.0 | 7.0 | 55.0 | 38.0 | 11.0 | 14.0 | 25.0 | 22.0 | 39.0 | 12.0 |
| 14 | 47.0 | 80.0 | 29.0 | 15.0 | 24.0 | 44.0 | 6.0 | 17.0 | 25.0 | 54.0 | 14.0 | 4.0 | 1.0 | 20.0 | 12.0 | 10.0 |
| 15 | 11.0 | 46.0 | 28.0 | 12.0 | 43.0 | 59.0 | 17.0 | 79.0 | 6.0 | 28.0 | 16.0 | 11.0 | 7.0 | 15.0 | 17.0 | 7.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -1.113 | -1.386 | -0.224 | -0.275 | -0.087 | -0.176 | -0.045 | 1.34 | -1.386 | -0.328 | 0.681 | 0.299 | -1.241 | -1.557 | 0.619 | -0.384 |
| 02 | -2.92 | -4.22 | 0.484 | -2.374 | -1.763 | -2.92 | 0.554 | -2.92 | -1.763 | -4.22 | 1.677 | -2.92 | -2.022 | -4.22 | 1.857 | -2.92 |
| 03 | -4.22 | -2.92 | -1.763 | -1.241 | -4.22 | -4.22 | -4.22 | -2.92 | -0.087 | 0.661 | 0.239 | 2.397 | -4.22 | -2.022 | -4.22 | -2.374 |
| 04 | -2.022 | -2.374 | -2.374 | -0.443 | -0.899 | -2.92 | -2.374 | 0.46 | -0.807 | -1.241 | -2.92 | -0.384 | 0.576 | 0.508 | -0.443 | 1.994 |
| 05 | -0.899 | -2.92 | 0.382 | -0.176 | -1.386 | -4.22 | -1.001 | 0.382 | -4.22 | -2.92 | -0.899 | -0.899 | -0.275 | -2.92 | 2.01 | 0.681 |
| 06 | -4.22 | 0.327 | -4.22 | -4.22 | -4.22 | -2.022 | -4.22 | -4.22 | -2.374 | 2.205 | -1.763 | -1.001 | -4.22 | 1.501 | -2.92 | -2.022 |
| 07 | -4.22 | -4.22 | -2.92 | -2.92 | -2.92 | 2.192 | -4.22 | 1.801 | -4.22 | -2.022 | -2.92 | -2.92 | -4.22 | -1.386 | -2.92 | -1.557 |
| 08 | -4.22 | -4.22 | -4.22 | -2.92 | 1.39 | 0.757 | -0.224 | 0.878 | -2.022 | -4.22 | -4.22 | -4.22 | 0.862 | -0.087 | 0.972 | -0.807 |
| 09 | 0.269 | -2.92 | 1.11 | 0.757 | -0.176 | -1.241 | -0.328 | 0.207 | -0.899 | -2.92 | 0.926 | -0.723 | 0.071 | -1.763 | 0.327 | -1.241 |
| 10 | -2.92 | -4.22 | 1.254 | -2.92 | -4.22 | -4.22 | -0.723 | -2.92 | -2.022 | -2.022 | 1.994 | -2.92 | -1.557 | -4.22 | 1.36 | -4.22 |
| 11 | -4.22 | -4.22 | -1.113 | -2.92 | -4.22 | -2.92 | -2.92 | -2.92 | 1.11 | 1.097 | 1.748 | 1.243 | -4.22 | -2.92 | -2.374 | -4.22 |
| 12 | -1.763 | -0.275 | 0.508 | -0.723 | 0.175 | -0.807 | -0.899 | 0.071 | 0.681 | 0.531 | 0.269 | 0.207 | -0.224 | -1.001 | 0.269 | 0.107 |
| 13 | 0.382 | -0.384 | 0.46 | -0.899 | 0.757 | -0.573 | -1.241 | -1.241 | 0.775 | 0.409 | -0.807 | -0.573 | -0.005 | -0.131 | 0.434 | -0.723 |
| 14 | 0.619 | 1.148 | 0.141 | -0.506 | -0.045 | 0.554 | -1.386 | -0.384 | -0.005 | 0.757 | -0.573 | -1.763 | -2.92 | -0.224 | -0.723 | -0.899 |
| 15 | -0.807 | 0.598 | 0.107 | -0.723 | 0.531 | 0.845 | -0.384 | 1.136 | -1.386 | 0.107 | -0.443 | -0.807 | -1.241 | -0.506 | -0.384 | -1.241 |