Model info
| Transcription factor | Runx3 | ||||||||
| Model | RUNX3_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 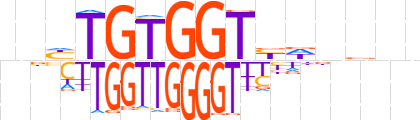 | ||||||||
| LOGO (reverse complement) | 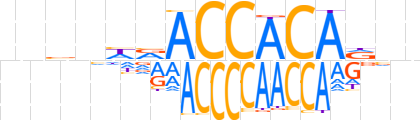 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 15 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | bnhTGTGGTbdnbvn | ||||||||
| Best auROC (human) | 0.994 | ||||||||
| Best auROC (mouse) | 0.979 | ||||||||
| Peak sets in benchmark (human) | 2 | ||||||||
| Peak sets in benchmark (mouse) | 27 | ||||||||
| Aligned words | 198 | ||||||||
| TF family | Runt-related factors {6.4.1} | ||||||||
| TF subfamily | Runx3 (PEBP2alphaC, CBF-alpha3, AML-2) {6.4.1.0.3} | ||||||||
| MGI | MGI:102672 | ||||||||
| EntrezGene | |||||||||
| UniProt ID | RUNX3_MOUSE | ||||||||
| UniProt AC | Q64131 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Runx3 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.0 | 6.0 | 12.0 | 1.0 | 10.0 | 8.0 | 6.0 | 17.0 | 6.0 | 16.0 | 31.0 | 25.0 | 8.0 | 14.0 | 15.0 | 9.0 |
| 02 | 3.0 | 13.0 | 1.0 | 7.0 | 7.0 | 28.0 | 2.0 | 7.0 | 20.0 | 38.0 | 4.0 | 2.0 | 3.0 | 25.0 | 5.0 | 19.0 |
| 03 | 5.0 | 0.0 | 0.0 | 28.0 | 2.0 | 0.0 | 0.0 | 102.0 | 1.0 | 0.0 | 0.0 | 11.0 | 3.0 | 1.0 | 0.0 | 31.0 |
| 04 | 0.0 | 0.0 | 11.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.0 | 0.0 | 168.0 | 1.0 |
| 05 | 0.0 | 1.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 8.0 | 7.0 | 1.0 | 164.0 | 0.0 | 0.0 | 0.0 | 1.0 |
| 06 | 0.0 | 0.0 | 8.0 | 0.0 | 0.0 | 0.0 | 8.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 166.0 | 0.0 |
| 07 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 182.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 08 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 2.0 | 8.0 | 173.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 09 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 3.0 | 0.0 | 5.0 | 12.0 | 60.0 | 17.0 | 85.0 |
| 10 | 8.0 | 1.0 | 0.0 | 3.0 | 26.0 | 0.0 | 1.0 | 37.0 | 10.0 | 0.0 | 4.0 | 3.0 | 28.0 | 4.0 | 23.0 | 36.0 |
| 11 | 17.0 | 18.0 | 29.0 | 8.0 | 1.0 | 1.0 | 1.0 | 2.0 | 5.0 | 6.0 | 5.0 | 12.0 | 9.0 | 13.0 | 32.0 | 25.0 |
| 12 | 7.0 | 10.0 | 14.0 | 1.0 | 6.0 | 18.0 | 4.0 | 10.0 | 10.0 | 26.0 | 16.0 | 15.0 | 0.0 | 7.0 | 30.0 | 10.0 |
| 13 | 2.0 | 9.0 | 10.0 | 2.0 | 19.0 | 26.0 | 4.0 | 12.0 | 15.0 | 18.0 | 23.0 | 8.0 | 0.0 | 7.0 | 25.0 | 4.0 |
| 14 | 3.0 | 9.0 | 14.0 | 10.0 | 19.0 | 14.0 | 7.0 | 20.0 | 2.0 | 12.0 | 28.0 | 20.0 | 1.0 | 8.0 | 15.0 | 2.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -3.591 | -0.626 | 0.041 | -2.188 | -0.136 | -0.351 | -0.626 | 0.382 | -0.626 | 0.322 | 0.974 | 0.762 | -0.351 | 0.192 | 0.259 | -0.237 |
| 02 | -1.269 | 0.119 | -2.188 | -0.479 | -0.479 | 0.873 | -1.626 | -0.479 | 0.542 | 1.176 | -1.006 | -1.626 | -1.269 | 0.762 | -0.798 | 0.491 |
| 03 | -0.798 | -3.591 | -3.591 | 0.873 | -1.626 | -3.591 | -3.591 | 2.158 | -2.188 | -3.591 | -3.591 | -0.043 | -1.269 | -2.188 | -3.591 | 0.974 |
| 04 | -3.591 | -3.591 | -0.043 | -3.591 | -3.591 | -3.591 | -2.188 | -3.591 | -3.591 | -3.591 | -3.591 | -3.591 | -1.269 | -3.591 | 2.656 | -2.188 |
| 05 | -3.591 | -2.188 | -3.591 | -1.626 | -3.591 | -3.591 | -3.591 | -3.591 | -0.351 | -0.479 | -2.188 | 2.632 | -3.591 | -3.591 | -3.591 | -2.188 |
| 06 | -3.591 | -3.591 | -0.351 | -3.591 | -3.591 | -3.591 | -0.351 | -3.591 | -3.591 | -3.591 | -2.188 | -3.591 | -3.591 | -2.188 | 2.644 | -3.591 |
| 07 | -3.591 | -3.591 | -3.591 | -3.591 | -3.591 | -3.591 | -2.188 | -3.591 | -3.591 | -2.188 | 2.736 | -3.591 | -3.591 | -3.591 | -3.591 | -3.591 |
| 08 | -3.591 | -3.591 | -3.591 | -3.591 | -3.591 | -3.591 | -3.591 | -2.188 | -3.591 | -1.626 | -0.351 | 2.685 | -3.591 | -3.591 | -3.591 | -3.591 |
| 09 | -3.591 | -3.591 | -3.591 | -3.591 | -3.591 | -2.188 | -3.591 | -2.188 | -3.591 | -1.269 | -3.591 | -0.798 | 0.041 | 1.629 | 0.382 | 1.976 |
| 10 | -0.351 | -2.188 | -3.591 | -1.269 | 0.8 | -3.591 | -2.188 | 1.149 | -0.136 | -3.591 | -1.006 | -1.269 | 0.873 | -1.006 | 0.679 | 1.122 |
| 11 | 0.382 | 0.438 | 0.908 | -0.351 | -2.188 | -2.188 | -2.188 | -1.626 | -0.798 | -0.626 | -0.798 | 0.041 | -0.237 | 0.119 | 1.006 | 0.762 |
| 12 | -0.479 | -0.136 | 0.192 | -2.188 | -0.626 | 0.438 | -1.006 | -0.136 | -0.136 | 0.8 | 0.322 | 0.259 | -3.591 | -0.479 | 0.942 | -0.136 |
| 13 | -1.626 | -0.237 | -0.136 | -1.626 | 0.491 | 0.8 | -1.006 | 0.041 | 0.259 | 0.438 | 0.679 | -0.351 | -3.591 | -0.479 | 0.762 | -1.006 |
| 14 | -1.269 | -0.237 | 0.192 | -0.136 | 0.491 | 0.192 | -0.479 | 0.542 | -1.626 | 0.041 | 0.873 | 0.542 | -2.188 | -0.351 | 0.259 | -1.626 |