Model info
| Transcription factor | SIX2 (GeneCards) | ||||||||
| Model | SIX2_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 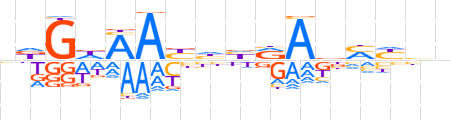 | ||||||||
| LOGO (reverse complement) | 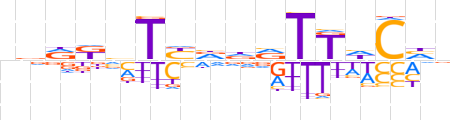 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 16 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | ndGdAAbhhRAbMYbn | ||||||||
| Best auROC (human) | 0.975 | ||||||||
| Best auROC (mouse) | 0.96 | ||||||||
| Peak sets in benchmark (human) | 18 | ||||||||
| Peak sets in benchmark (mouse) | 9 | ||||||||
| Aligned words | 489 | ||||||||
| TF family | HD-SINE factors {3.1.6} | ||||||||
| TF subfamily | SIX1-like factors {3.1.6.1} | ||||||||
| HGNC | HGNC:10888 | ||||||||
| EntrezGene | GeneID:10736 (SSTAR profile) | ||||||||
| UniProt ID | SIX2_HUMAN | ||||||||
| UniProt AC | Q9NPC8 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | SIX2 expression | ||||||||
| ReMap ChIP-seq dataset list | SIX2 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 34.0 | 1.0 | 45.0 | 37.0 | 40.0 | 5.0 | 7.0 | 90.0 | 26.0 | 0.0 | 33.0 | 55.0 | 11.0 | 4.0 | 49.0 | 44.0 |
| 02 | 1.0 | 6.0 | 100.0 | 4.0 | 1.0 | 0.0 | 6.0 | 3.0 | 6.0 | 4.0 | 120.0 | 4.0 | 5.0 | 3.0 | 210.0 | 8.0 |
| 03 | 4.0 | 3.0 | 5.0 | 1.0 | 7.0 | 2.0 | 1.0 | 3.0 | 190.0 | 25.0 | 45.0 | 176.0 | 5.0 | 1.0 | 6.0 | 7.0 |
| 04 | 171.0 | 16.0 | 6.0 | 13.0 | 24.0 | 5.0 | 1.0 | 1.0 | 45.0 | 8.0 | 1.0 | 3.0 | 148.0 | 14.0 | 9.0 | 16.0 |
| 05 | 374.0 | 2.0 | 6.0 | 6.0 | 41.0 | 0.0 | 1.0 | 1.0 | 13.0 | 2.0 | 1.0 | 1.0 | 27.0 | 3.0 | 2.0 | 1.0 |
| 06 | 31.0 | 223.0 | 41.0 | 160.0 | 1.0 | 4.0 | 0.0 | 2.0 | 2.0 | 5.0 | 0.0 | 3.0 | 0.0 | 4.0 | 3.0 | 2.0 |
| 07 | 9.0 | 16.0 | 2.0 | 7.0 | 45.0 | 114.0 | 5.0 | 72.0 | 11.0 | 21.0 | 2.0 | 10.0 | 35.0 | 49.0 | 16.0 | 67.0 |
| 08 | 27.0 | 12.0 | 15.0 | 46.0 | 22.0 | 42.0 | 9.0 | 127.0 | 8.0 | 1.0 | 4.0 | 12.0 | 20.0 | 31.0 | 27.0 | 78.0 |
| 09 | 24.0 | 3.0 | 43.0 | 7.0 | 31.0 | 12.0 | 28.0 | 15.0 | 9.0 | 7.0 | 32.0 | 7.0 | 30.0 | 28.0 | 187.0 | 18.0 |
| 10 | 80.0 | 5.0 | 5.0 | 4.0 | 42.0 | 5.0 | 1.0 | 2.0 | 279.0 | 4.0 | 4.0 | 3.0 | 40.0 | 4.0 | 3.0 | 0.0 |
| 11 | 49.0 | 87.0 | 158.0 | 147.0 | 5.0 | 7.0 | 3.0 | 3.0 | 2.0 | 5.0 | 4.0 | 2.0 | 1.0 | 4.0 | 4.0 | 0.0 |
| 12 | 28.0 | 23.0 | 2.0 | 4.0 | 63.0 | 28.0 | 0.0 | 12.0 | 76.0 | 68.0 | 3.0 | 22.0 | 57.0 | 81.0 | 7.0 | 7.0 |
| 13 | 36.0 | 131.0 | 21.0 | 36.0 | 19.0 | 109.0 | 5.0 | 67.0 | 1.0 | 5.0 | 1.0 | 5.0 | 5.0 | 26.0 | 2.0 | 12.0 |
| 14 | 10.0 | 18.0 | 28.0 | 5.0 | 49.0 | 123.0 | 29.0 | 70.0 | 0.0 | 15.0 | 7.0 | 7.0 | 10.0 | 45.0 | 39.0 | 26.0 |
| 15 | 13.0 | 20.0 | 27.0 | 9.0 | 85.0 | 47.0 | 14.0 | 55.0 | 16.0 | 23.0 | 46.0 | 18.0 | 17.0 | 26.0 | 30.0 | 35.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.122 | -3.09 | 0.399 | 0.205 | 0.282 | -1.732 | -1.416 | 1.088 | -0.143 | -4.368 | 0.092 | 0.598 | -0.984 | -1.938 | 0.484 | 0.377 |
| 02 | -3.09 | -1.562 | 1.193 | -1.938 | -3.09 | -4.368 | -1.562 | -2.196 | -1.562 | -1.938 | 1.375 | -1.938 | -1.732 | -2.196 | 1.933 | -1.289 |
| 03 | -1.938 | -2.196 | -1.732 | -3.09 | -1.416 | -2.546 | -3.09 | -2.196 | 1.833 | -0.182 | 0.399 | 1.757 | -1.732 | -3.09 | -1.562 | -1.416 |
| 04 | 1.728 | -0.62 | -1.562 | -0.822 | -0.222 | -1.732 | -3.09 | -3.09 | 0.399 | -1.289 | -3.09 | -2.196 | 1.584 | -0.75 | -1.177 | -0.62 |
| 05 | 2.509 | -2.546 | -1.562 | -1.562 | 0.307 | -4.368 | -3.09 | -3.09 | -0.822 | -2.546 | -3.09 | -3.09 | -0.106 | -2.196 | -2.546 | -3.09 |
| 06 | 0.03 | 1.993 | 0.307 | 1.662 | -3.09 | -1.938 | -4.368 | -2.546 | -2.546 | -1.732 | -4.368 | -2.196 | -4.368 | -1.938 | -2.196 | -2.546 |
| 07 | -1.177 | -0.62 | -2.546 | -1.416 | 0.399 | 1.324 | -1.732 | 0.866 | -0.984 | -0.353 | -2.546 | -1.076 | 0.15 | 0.484 | -0.62 | 0.794 |
| 08 | -0.106 | -0.899 | -0.683 | 0.421 | -0.308 | 0.331 | -1.177 | 1.431 | -1.289 | -3.09 | -1.938 | -0.899 | -0.401 | 0.03 | -0.106 | 0.946 |
| 09 | -0.222 | -2.196 | 0.354 | -1.416 | 0.03 | -0.899 | -0.07 | -0.683 | -1.177 | -1.416 | 0.062 | -1.416 | -0.002 | -0.07 | 1.817 | -0.504 |
| 10 | 0.971 | -1.732 | -1.732 | -1.938 | 0.331 | -1.732 | -3.09 | -2.546 | 2.217 | -1.938 | -1.938 | -2.196 | 0.282 | -1.938 | -2.196 | -4.368 |
| 11 | 0.484 | 1.054 | 1.649 | 1.577 | -1.732 | -1.416 | -2.196 | -2.196 | -2.546 | -1.732 | -1.938 | -2.546 | -3.09 | -1.938 | -1.938 | -4.368 |
| 12 | -0.07 | -0.264 | -2.546 | -1.938 | 0.733 | -0.07 | -4.368 | -0.899 | 0.92 | 0.809 | -2.196 | -0.308 | 0.634 | 0.983 | -1.416 | -1.416 |
| 13 | 0.178 | 1.462 | -0.353 | 0.178 | -0.451 | 1.279 | -1.732 | 0.794 | -3.09 | -1.732 | -3.09 | -1.732 | -1.732 | -0.143 | -2.546 | -0.899 |
| 14 | -1.076 | -0.504 | -0.07 | -1.732 | 0.484 | 1.399 | -0.036 | 0.838 | -4.368 | -0.683 | -1.416 | -1.416 | -1.076 | 0.399 | 0.257 | -0.143 |
| 15 | -0.822 | -0.401 | -0.106 | -1.177 | 1.031 | 0.442 | -0.75 | 0.598 | -0.62 | -0.264 | 0.421 | -0.504 | -0.56 | -0.143 | -0.002 | 0.15 |