Model info
| Transcription factor | Six4 | ||||||||
| Model | SIX4_MOUSE.H11MO.0.C | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 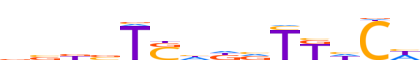 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 14 | ||||||||
| Quality | C | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | dGdMAbhdSAvhbb | ||||||||
| Best auROC (human) | |||||||||
| Best auROC (mouse) | 0.912 | ||||||||
| Peak sets in benchmark (human) | |||||||||
| Peak sets in benchmark (mouse) | 4 | ||||||||
| Aligned words | 487 | ||||||||
| TF family | HD-SINE factors {3.1.6} | ||||||||
| TF subfamily | SIX4-like factors {3.1.6.3} | ||||||||
| MGI | MGI:106034 | ||||||||
| EntrezGene | GeneID:20474 (SSTAR profile) | ||||||||
| UniProt ID | SIX4_MOUSE | ||||||||
| UniProt AC | Q61321 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Six4 expression | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 166.0 | 20.0 | 78.0 | 223.0 |
| 02 | 16.0 | 13.0 | 444.0 | 14.0 |
| 03 | 235.0 | 33.0 | 90.0 | 129.0 |
| 04 | 348.0 | 77.0 | 41.0 | 21.0 |
| 05 | 410.0 | 5.0 | 53.0 | 19.0 |
| 06 | 28.0 | 236.0 | 109.0 | 114.0 |
| 07 | 112.0 | 251.0 | 31.0 | 93.0 |
| 08 | 96.0 | 68.0 | 89.0 | 234.0 |
| 09 | 57.0 | 94.0 | 323.0 | 13.0 |
| 10 | 426.0 | 14.0 | 18.0 | 29.0 |
| 11 | 56.0 | 166.0 | 212.0 | 53.0 |
| 12 | 240.0 | 124.0 | 61.0 | 62.0 |
| 13 | 61.0 | 232.0 | 126.0 | 68.0 |
| 14 | 75.0 | 198.0 | 98.0 | 116.0 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.307 | -1.744 | -0.438 | 0.599 |
| 02 | -1.95 | -2.137 | 1.285 | -2.071 |
| 03 | 0.652 | -1.272 | -0.298 | 0.057 |
| 04 | 1.042 | -0.451 | -1.064 | -1.699 |
| 05 | 1.205 | -2.936 | -0.816 | -1.792 |
| 06 | -1.429 | 0.656 | -0.109 | -0.065 |
| 07 | -0.082 | 0.717 | -1.332 | -0.265 |
| 08 | -0.234 | -0.573 | -0.309 | 0.647 |
| 09 | -0.745 | -0.255 | 0.968 | -2.137 |
| 10 | 1.243 | -2.071 | -1.842 | -1.395 |
| 11 | -0.762 | 0.307 | 0.549 | -0.816 |
| 12 | 0.672 | 0.018 | -0.679 | -0.663 |
| 13 | -0.679 | 0.639 | 0.034 | -0.573 |
| 14 | -0.477 | 0.481 | -0.214 | -0.048 |