Model info
| Transcription factor | SMARCA5 (GeneCards) | ||||||||
| Model | SMCA5_HUMAN.H11DI.0.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 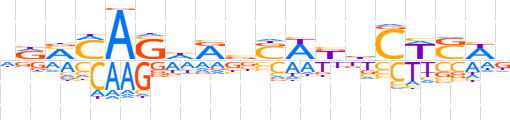 | ||||||||
| LOGO (reverse complement) | 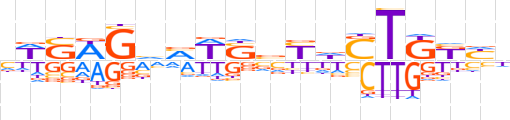 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 18 | ||||||||
| Quality | C | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | dRMMAShRbSWKbCWSWd | ||||||||
| Best auROC (human) | 0.668 | ||||||||
| Best auROC (mouse) | 0.729 | ||||||||
| Peak sets in benchmark (human) | 3 | ||||||||
| Peak sets in benchmark (mouse) | 2 | ||||||||
| Aligned words | 274 | ||||||||
| TF family | Myb/SANT domain factors {3.5.1} | ||||||||
| TF subfamily | SMARCA-like factors {3.5.1.5} | ||||||||
| HGNC | HGNC:11101 | ||||||||
| EntrezGene | GeneID:8467 (SSTAR profile) | ||||||||
| UniProt ID | SMCA5_HUMAN | ||||||||
| UniProt AC | O60264 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | SMARCA5 expression | ||||||||
| ReMap ChIP-seq dataset list | SMARCA5 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 8.0 | 3.0 | 95.0 | 14.0 | 8.0 | 7.0 | 13.0 | 12.0 | 14.0 | 6.0 | 22.0 | 3.0 | 11.0 | 9.0 | 30.0 | 9.0 |
| 02 | 22.0 | 12.0 | 4.0 | 3.0 | 16.0 | 2.0 | 5.0 | 2.0 | 108.0 | 29.0 | 11.0 | 12.0 | 23.0 | 6.0 | 8.0 | 1.0 |
| 03 | 23.0 | 128.0 | 3.0 | 15.0 | 5.0 | 37.0 | 2.0 | 5.0 | 6.0 | 19.0 | 2.0 | 1.0 | 4.0 | 13.0 | 0.0 | 1.0 |
| 04 | 38.0 | 0.0 | 0.0 | 0.0 | 190.0 | 0.0 | 5.0 | 2.0 | 5.0 | 0.0 | 2.0 | 0.0 | 19.0 | 0.0 | 3.0 | 0.0 |
| 05 | 15.0 | 29.0 | 194.0 | 14.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 2.0 | 6.0 | 1.0 | 0.0 | 0.0 | 2.0 | 0.0 |
| 06 | 13.0 | 1.0 | 1.0 | 1.0 | 12.0 | 8.0 | 1.0 | 10.0 | 110.0 | 23.0 | 27.0 | 42.0 | 2.0 | 9.0 | 3.0 | 1.0 |
| 07 | 98.0 | 15.0 | 21.0 | 3.0 | 29.0 | 7.0 | 3.0 | 2.0 | 17.0 | 2.0 | 13.0 | 0.0 | 28.0 | 4.0 | 22.0 | 0.0 |
| 08 | 6.0 | 32.0 | 100.0 | 34.0 | 14.0 | 11.0 | 1.0 | 2.0 | 6.0 | 34.0 | 11.0 | 8.0 | 1.0 | 1.0 | 2.0 | 1.0 |
| 09 | 2.0 | 13.0 | 3.0 | 9.0 | 7.0 | 60.0 | 5.0 | 6.0 | 14.0 | 76.0 | 18.0 | 6.0 | 0.0 | 33.0 | 10.0 | 2.0 |
| 10 | 22.0 | 1.0 | 0.0 | 0.0 | 130.0 | 14.0 | 3.0 | 35.0 | 16.0 | 7.0 | 0.0 | 13.0 | 12.0 | 3.0 | 2.0 | 6.0 |
| 11 | 7.0 | 13.0 | 33.0 | 127.0 | 4.0 | 6.0 | 0.0 | 15.0 | 0.0 | 2.0 | 1.0 | 2.0 | 7.0 | 18.0 | 13.0 | 16.0 |
| 12 | 4.0 | 3.0 | 5.0 | 6.0 | 11.0 | 17.0 | 1.0 | 10.0 | 15.0 | 8.0 | 14.0 | 10.0 | 8.0 | 15.0 | 43.0 | 94.0 |
| 13 | 2.0 | 31.0 | 4.0 | 1.0 | 5.0 | 33.0 | 4.0 | 1.0 | 3.0 | 52.0 | 7.0 | 1.0 | 4.0 | 108.0 | 6.0 | 2.0 |
| 14 | 1.0 | 4.0 | 0.0 | 9.0 | 34.0 | 21.0 | 2.0 | 167.0 | 3.0 | 12.0 | 3.0 | 3.0 | 1.0 | 0.0 | 1.0 | 3.0 |
| 15 | 2.0 | 23.0 | 14.0 | 0.0 | 3.0 | 31.0 | 3.0 | 0.0 | 2.0 | 1.0 | 3.0 | 0.0 | 6.0 | 113.0 | 58.0 | 5.0 |
| 16 | 6.0 | 2.0 | 1.0 | 4.0 | 121.0 | 25.0 | 1.0 | 21.0 | 33.0 | 7.0 | 4.0 | 34.0 | 2.0 | 1.0 | 0.0 | 2.0 |
| 17 | 22.0 | 25.0 | 102.0 | 13.0 | 10.0 | 12.0 | 4.0 | 9.0 | 0.0 | 3.0 | 2.0 | 1.0 | 14.0 | 1.0 | 15.0 | 31.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.702 | -1.616 | 1.733 | -0.161 | -0.702 | -0.83 | -0.233 | -0.311 | -0.161 | -0.976 | 0.282 | -1.616 | -0.395 | -0.589 | 0.588 | -0.589 |
| 02 | 0.282 | -0.311 | -1.354 | -1.616 | -0.03 | -1.97 | -1.147 | -1.97 | 1.861 | 0.555 | -0.395 | -0.311 | 0.326 | -0.976 | -0.702 | -2.525 |
| 03 | 0.326 | 2.03 | -1.616 | -0.093 | -1.147 | 0.796 | -1.97 | -1.147 | -0.976 | 0.138 | -1.97 | -2.525 | -1.354 | -0.233 | -3.878 | -2.525 |
| 04 | 0.822 | -3.878 | -3.878 | -3.878 | 2.425 | -3.878 | -1.147 | -1.97 | -1.147 | -3.878 | -1.97 | -3.878 | 0.138 | -3.878 | -1.616 | -3.878 |
| 05 | -0.093 | 0.555 | 2.445 | -0.161 | -3.878 | -3.878 | -3.878 | -3.878 | -2.525 | -1.97 | -0.976 | -2.525 | -3.878 | -3.878 | -1.97 | -3.878 |
| 06 | -0.233 | -2.525 | -2.525 | -2.525 | -0.311 | -0.702 | -2.525 | -0.487 | 1.879 | 0.326 | 0.484 | 0.922 | -1.97 | -0.589 | -1.616 | -2.525 |
| 07 | 1.764 | -0.093 | 0.237 | -1.616 | 0.555 | -0.83 | -1.616 | -1.97 | 0.029 | -1.97 | -0.233 | -3.878 | 0.52 | -1.354 | 0.282 | -3.878 |
| 08 | -0.976 | 0.652 | 1.784 | 0.712 | -0.161 | -0.395 | -2.525 | -1.97 | -0.976 | 0.712 | -0.395 | -0.702 | -2.525 | -2.525 | -1.97 | -2.525 |
| 09 | -1.97 | -0.233 | -1.616 | -0.589 | -0.83 | 1.276 | -1.147 | -0.976 | -0.161 | 1.511 | 0.085 | -0.976 | -3.878 | 0.683 | -0.487 | -1.97 |
| 10 | 0.282 | -2.525 | -3.878 | -3.878 | 2.046 | -0.161 | -1.616 | 0.741 | -0.03 | -0.83 | -3.878 | -0.233 | -0.311 | -1.616 | -1.97 | -0.976 |
| 11 | -0.83 | -0.233 | 0.683 | 2.023 | -1.354 | -0.976 | -3.878 | -0.093 | -3.878 | -1.97 | -2.525 | -1.97 | -0.83 | 0.085 | -0.233 | -0.03 |
| 12 | -1.354 | -1.616 | -1.147 | -0.976 | -0.395 | 0.029 | -2.525 | -0.487 | -0.093 | -0.702 | -0.161 | -0.487 | -0.702 | -0.093 | 0.945 | 1.723 |
| 13 | -1.97 | 0.621 | -1.354 | -2.525 | -1.147 | 0.683 | -1.354 | -2.525 | -1.616 | 1.134 | -0.83 | -2.525 | -1.354 | 1.861 | -0.976 | -1.97 |
| 14 | -2.525 | -1.354 | -3.878 | -0.589 | 0.712 | 0.237 | -1.97 | 2.296 | -1.616 | -0.311 | -1.616 | -1.616 | -2.525 | -3.878 | -2.525 | -1.616 |
| 15 | -1.97 | 0.326 | -0.161 | -3.878 | -1.616 | 0.621 | -1.616 | -3.878 | -1.97 | -2.525 | -1.616 | -3.878 | -0.976 | 1.906 | 1.242 | -1.147 |
| 16 | -0.976 | -1.97 | -2.525 | -1.354 | 1.974 | 0.408 | -2.525 | 0.237 | 0.683 | -0.83 | -1.354 | 0.712 | -1.97 | -2.525 | -3.878 | -1.97 |
| 17 | 0.282 | 0.408 | 1.804 | -0.233 | -0.487 | -0.311 | -1.354 | -0.589 | -3.878 | -1.616 | -1.97 | -2.525 | -0.161 | -2.525 | -0.093 | 0.621 |