Model info
| Transcription factor | Snai2 | ||||||||
| Model | SNAI2_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 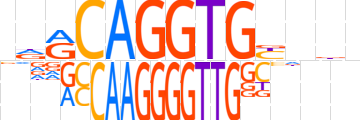 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 13 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | ndRCAGGTGYnbb | ||||||||
| Best auROC (human) | 0.976 | ||||||||
| Best auROC (mouse) | 0.94 | ||||||||
| Peak sets in benchmark (human) | 11 | ||||||||
| Peak sets in benchmark (mouse) | 3 | ||||||||
| Aligned words | 417 | ||||||||
| TF family | More than 3 adjacent zinc finger factors {2.3.3} | ||||||||
| TF subfamily | Snail-like factors {2.3.3.2} | ||||||||
| MGI | MGI:1096393 | ||||||||
| EntrezGene | GeneID:20583 (SSTAR profile) | ||||||||
| UniProt ID | SNAI2_MOUSE | ||||||||
| UniProt AC | P97469 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Snai2 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 27.0 | 1.0 | 42.0 | 4.0 | 65.0 | 3.0 | 52.0 | 25.0 | 56.0 | 17.0 | 34.0 | 11.0 | 18.0 | 4.0 | 46.0 | 4.0 |
| 02 | 73.0 | 1.0 | 92.0 | 0.0 | 14.0 | 0.0 | 11.0 | 0.0 | 74.0 | 1.0 | 99.0 | 0.0 | 15.0 | 0.0 | 29.0 | 0.0 |
| 03 | 0.0 | 176.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 231.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 04 | 0.0 | 0.0 | 0.0 | 0.0 | 409.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 05 | 0.0 | 0.0 | 409.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 06 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 409.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 07 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 409.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 08 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 407.0 | 0.0 |
| 09 | 0.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 9.0 | 226.0 | 79.0 | 93.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 10 | 2.0 | 2.0 | 2.0 | 3.0 | 84.0 | 37.0 | 54.0 | 52.0 | 45.0 | 18.0 | 12.0 | 5.0 | 10.0 | 28.0 | 29.0 | 26.0 |
| 11 | 16.0 | 31.0 | 50.0 | 44.0 | 11.0 | 14.0 | 15.0 | 45.0 | 8.0 | 32.0 | 32.0 | 25.0 | 8.0 | 16.0 | 29.0 | 33.0 |
| 12 | 10.0 | 7.0 | 22.0 | 4.0 | 27.0 | 30.0 | 18.0 | 18.0 | 18.0 | 45.0 | 51.0 | 12.0 | 9.0 | 52.0 | 55.0 | 31.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.054 | -2.937 | 0.491 | -1.78 | 0.924 | -2.039 | 0.703 | -0.022 | 0.776 | -0.401 | 0.282 | -0.824 | -0.345 | -1.78 | 0.581 | -1.78 |
| 02 | 1.04 | -2.937 | 1.27 | -4.234 | -0.59 | -4.234 | -0.824 | -4.234 | 1.053 | -2.937 | 1.343 | -4.234 | -0.523 | -4.234 | 0.124 | -4.234 |
| 03 | -4.234 | 1.917 | -4.234 | -4.234 | -4.234 | -2.39 | -4.234 | -4.234 | -4.234 | 2.188 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 |
| 04 | -4.234 | -4.234 | -4.234 | -4.234 | 2.759 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 |
| 05 | -4.234 | -4.234 | 2.759 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 |
| 06 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | 2.759 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 |
| 07 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | 2.759 | -4.234 | -4.234 | -4.234 | -4.234 |
| 08 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | -2.39 | -4.234 | 2.754 | -4.234 |
| 09 | -4.234 | -2.937 | -2.937 | -4.234 | -4.234 | -4.234 | -4.234 | -4.234 | -1.018 | 2.166 | 1.118 | 1.281 | -4.234 | -4.234 | -4.234 | -4.234 |
| 10 | -2.39 | -2.39 | -2.39 | -2.039 | 1.18 | 0.365 | 0.74 | 0.703 | 0.559 | -0.345 | -0.74 | -1.574 | -0.916 | 0.09 | 0.124 | 0.017 |
| 11 | -0.46 | 0.19 | 0.664 | 0.537 | -0.824 | -0.59 | -0.523 | 0.559 | -1.13 | 0.222 | 0.222 | -0.022 | -1.13 | -0.46 | 0.124 | 0.252 |
| 12 | -0.916 | -1.258 | -0.148 | -1.78 | 0.054 | 0.158 | -0.345 | -0.345 | -0.345 | 0.559 | 0.683 | -0.74 | -1.018 | 0.703 | 0.758 | 0.19 |