Model info
| Transcription factor | SOX10 (GeneCards) | ||||||||
| Model | SOX10_HUMAN.H11MO.0.B | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO | 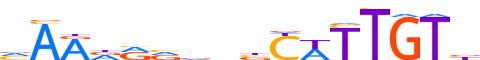 | ||||||||
| LOGO (reverse complement) | 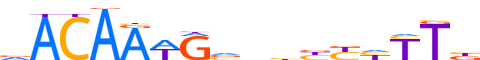 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 16 | ||||||||
| Quality | B | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | vARdRvvnbYWTTGTb | ||||||||
| Best auROC (human) | 0.91 | ||||||||
| Best auROC (mouse) | 0.763 | ||||||||
| Peak sets in benchmark (human) | 3 | ||||||||
| Peak sets in benchmark (mouse) | 3 | ||||||||
| Aligned words | 500 | ||||||||
| TF family | SOX-related factors {4.1.1} | ||||||||
| TF subfamily | Group E {4.1.1.5} | ||||||||
| HGNC | HGNC:11190 | ||||||||
| EntrezGene | GeneID:6663 (SSTAR profile) | ||||||||
| UniProt ID | SOX10_HUMAN | ||||||||
| UniProt AC | P56693 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | SOX10 expression | ||||||||
| ReMap ChIP-seq dataset list | SOX10 datasets | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 190.0 | 201.0 | 84.0 | 25.0 |
| 02 | 444.0 | 32.0 | 16.0 | 8.0 |
| 03 | 385.0 | 16.0 | 60.0 | 39.0 |
| 04 | 240.0 | 16.0 | 125.0 | 119.0 |
| 05 | 162.0 | 39.0 | 273.0 | 26.0 |
| 06 | 139.0 | 82.0 | 251.0 | 28.0 |
| 07 | 184.0 | 106.0 | 152.0 | 58.0 |
| 08 | 146.0 | 74.0 | 161.0 | 119.0 |
| 09 | 25.0 | 146.0 | 202.0 | 127.0 |
| 10 | 25.0 | 399.0 | 23.0 | 53.0 |
| 11 | 253.0 | 22.0 | 6.0 | 219.0 |
| 12 | 14.0 | 37.0 | 18.0 | 431.0 |
| 13 | 3.0 | 11.0 | 3.0 | 483.0 |
| 14 | 27.0 | 10.0 | 463.0 | 0.0 |
| 15 | 31.0 | 1.0 | 6.0 | 462.0 |
| 16 | 69.0 | 91.0 | 90.0 | 250.0 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.415 | 0.47 | -0.392 | -1.561 |
| 02 | 1.259 | -1.328 | -1.975 | -2.584 |
| 03 | 1.117 | -1.975 | -0.721 | -1.138 |
| 04 | 0.646 | -1.975 | 0.0 | -0.049 |
| 05 | 0.256 | -1.138 | 0.774 | -1.525 |
| 06 | 0.105 | -0.415 | 0.691 | -1.454 |
| 07 | 0.383 | -0.163 | 0.193 | -0.754 |
| 08 | 0.154 | -0.516 | 0.25 | -0.049 |
| 09 | -1.561 | 0.154 | 0.475 | 0.016 |
| 10 | -1.561 | 1.152 | -1.64 | -0.841 |
| 11 | 0.699 | -1.681 | -2.819 | 0.555 |
| 12 | -2.096 | -1.189 | -1.868 | 1.229 |
| 13 | -3.325 | -2.311 | -3.325 | 1.343 |
| 14 | -1.489 | -2.394 | 1.3 | -4.4 |
| 15 | -1.358 | -3.903 | -2.819 | 1.298 |
| 16 | -0.584 | -0.313 | -0.324 | 0.687 |