Model info
| Transcription factor | Sp1 | ||||||||
| Model | SP1_MOUSE.H11MO.0.A | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO | 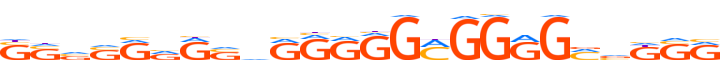 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 24 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | SSvSRvRSvRRRGGMGGRGMvKSR | ||||||||
| Best auROC (human) | 0.999 | ||||||||
| Best auROC (mouse) | 0.988 | ||||||||
| Peak sets in benchmark (human) | 53 | ||||||||
| Peak sets in benchmark (mouse) | 11 | ||||||||
| Aligned words | 489 | ||||||||
| TF family | Three-zinc finger Krüppel-related factors {2.3.1} | ||||||||
| TF subfamily | Sp1-like factors {2.3.1.1} | ||||||||
| MGI | MGI:98372 | ||||||||
| EntrezGene | GeneID:20683 (SSTAR profile) | ||||||||
| UniProt ID | SP1_MOUSE | ||||||||
| UniProt AC | O89090 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Sp1 expression | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 29.0 | 69.0 | 349.0 | 42.0 |
| 02 | 61.0 | 76.0 | 318.0 | 34.0 |
| 03 | 113.0 | 129.0 | 222.0 | 25.0 |
| 04 | 68.0 | 87.0 | 308.0 | 26.0 |
| 05 | 66.0 | 55.0 | 355.0 | 13.0 |
| 06 | 117.0 | 58.0 | 270.0 | 44.0 |
| 07 | 77.0 | 24.0 | 361.0 | 27.0 |
| 08 | 77.0 | 119.0 | 279.0 | 14.0 |
| 09 | 195.0 | 117.0 | 110.0 | 67.0 |
| 10 | 84.0 | 43.0 | 351.0 | 11.0 |
| 11 | 64.0 | 27.0 | 377.0 | 21.0 |
| 12 | 86.0 | 26.0 | 360.0 | 17.0 |
| 13 | 56.0 | 24.0 | 391.0 | 18.0 |
| 14 | 24.0 | 8.0 | 453.0 | 4.0 |
| 15 | 152.0 | 299.0 | 7.0 | 31.0 |
| 16 | 32.0 | 6.0 | 447.0 | 4.0 |
| 17 | 20.0 | 11.0 | 454.0 | 4.0 |
| 18 | 126.0 | 14.0 | 340.0 | 9.0 |
| 19 | 17.0 | 9.0 | 455.0 | 8.0 |
| 20 | 122.0 | 284.0 | 57.0 | 26.0 |
| 21 | 88.0 | 188.0 | 170.0 | 43.0 |
| 22 | 57.0 | 38.0 | 335.0 | 59.0 |
| 23 | 58.0 | 63.0 | 350.0 | 18.0 |
| 24 | 77.0 | 58.0 | 342.0 | 12.0 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -1.399 | -0.562 | 1.041 | -1.045 |
| 02 | -0.683 | -0.468 | 0.948 | -1.248 |
| 03 | -0.078 | 0.053 | 0.591 | -1.54 |
| 04 | -0.577 | -0.335 | 0.916 | -1.503 |
| 05 | -0.606 | -0.784 | 1.058 | -2.141 |
| 06 | -0.043 | -0.732 | 0.785 | -1.0 |
| 07 | -0.455 | -1.578 | 1.075 | -1.467 |
| 08 | -0.455 | -0.027 | 0.818 | -2.075 |
| 09 | 0.462 | -0.043 | -0.104 | -0.591 |
| 10 | -0.37 | -1.022 | 1.047 | -2.289 |
| 11 | -0.636 | -1.467 | 1.118 | -1.703 |
| 12 | -0.346 | -1.503 | 1.072 | -1.898 |
| 13 | -0.766 | -1.578 | 1.154 | -1.846 |
| 14 | -1.578 | -2.562 | 1.301 | -3.105 |
| 15 | 0.215 | 0.887 | -2.673 | -1.336 |
| 16 | -1.306 | -2.797 | 1.287 | -3.105 |
| 17 | -1.748 | -2.289 | 1.303 | -3.105 |
| 18 | 0.03 | -2.075 | 1.015 | -2.463 |
| 19 | -1.898 | -2.463 | 1.305 | -2.562 |
| 20 | -0.002 | 0.836 | -0.749 | -1.503 |
| 21 | -0.324 | 0.426 | 0.326 | -1.022 |
| 22 | -0.749 | -1.141 | 1.0 | -0.715 |
| 23 | -0.732 | -0.651 | 1.044 | -1.846 |
| 24 | -0.455 | -0.732 | 1.021 | -2.212 |