Model info
| Transcription factor | SP2 (GeneCards) | ||||||||
| Model | SP2_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 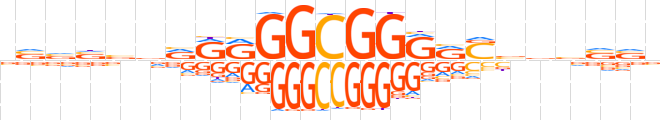 | ||||||||
| LOGO (reverse complement) | 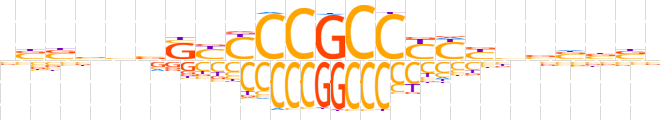 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv10 | ||||||||
| Model length | 23 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | vvvvbvRRRGGCGGRRSbdvvvb | ||||||||
| Best auROC (human) | 0.978 | ||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | 16 | ||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 501 | ||||||||
| TF family | Three-zinc finger Krüppel-related factors {2.3.1} | ||||||||
| TF subfamily | Sp1-like factors {2.3.1.1} | ||||||||
| HGNC | HGNC:11207 | ||||||||
| EntrezGene | GeneID:6668 (SSTAR profile) | ||||||||
| UniProt ID | SP2_HUMAN | ||||||||
| UniProt AC | Q02086 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | SP2 expression | ||||||||
| ReMap ChIP-seq dataset list | SP2 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 12.99 | 13.311 | 48.535 | 1.147 | 24.625 | 52.992 | 80.109 | 12.481 | 34.452 | 47.039 | 95.109 | 7.399 | 4.248 | 18.769 | 31.139 | 7.803 |
| 02 | 17.116 | 18.109 | 38.294 | 2.795 | 21.339 | 29.267 | 63.636 | 17.87 | 46.023 | 71.283 | 112.583 | 25.004 | 5.628 | 11.99 | 9.131 | 2.08 |
| 03 | 14.137 | 17.569 | 56.598 | 1.802 | 10.633 | 36.186 | 59.089 | 24.742 | 26.719 | 57.347 | 122.356 | 17.221 | 3.008 | 8.583 | 28.52 | 7.638 |
| 04 | 13.993 | 12.867 | 21.911 | 5.726 | 7.113 | 39.496 | 45.699 | 27.376 | 36.311 | 97.576 | 107.431 | 25.246 | 3.441 | 11.156 | 27.363 | 9.442 |
| 05 | 25.432 | 3.159 | 30.323 | 1.944 | 27.699 | 40.903 | 66.055 | 26.439 | 69.627 | 57.737 | 61.13 | 13.91 | 11.15 | 12.048 | 29.432 | 15.16 |
| 06 | 37.029 | 7.153 | 85.56 | 4.166 | 17.457 | 20.961 | 64.888 | 10.54 | 56.565 | 31.778 | 89.233 | 9.364 | 5.317 | 8.115 | 42.091 | 1.931 |
| 07 | 9.901 | 4.408 | 89.411 | 12.647 | 9.682 | 3.967 | 44.687 | 9.671 | 39.016 | 18.598 | 207.636 | 16.522 | 0.0 | 2.245 | 19.759 | 3.998 |
| 08 | 21.947 | 3.181 | 32.433 | 1.037 | 7.936 | 1.952 | 16.914 | 2.416 | 84.407 | 27.569 | 243.703 | 5.814 | 3.126 | 1.097 | 36.212 | 2.404 |
| 09 | 5.458 | 1.139 | 108.628 | 2.191 | 2.004 | 0.917 | 30.878 | 0.0 | 6.297 | 4.3 | 312.627 | 6.038 | 2.217 | 0.0 | 9.455 | 0.0 |
| 10 | 1.227 | 0.0 | 14.75 | 0.0 | 0.0 | 0.0 | 6.357 | 0.0 | 1.068 | 8.429 | 452.09 | 0.0 | 0.0 | 0.0 | 8.228 | 0.0 |
| 11 | 0.0 | 1.227 | 0.0 | 1.068 | 0.0 | 8.429 | 0.0 | 0.0 | 13.489 | 450.639 | 3.714 | 13.583 | 0.0 | 0.0 | 0.0 | 0.0 |
| 12 | 0.0 | 0.0 | 13.489 | 0.0 | 9.047 | 6.073 | 444.247 | 0.926 | 0.0 | 0.0 | 3.714 | 0.0 | 0.0 | 0.0 | 14.652 | 0.0 |
| 13 | 1.154 | 0.0 | 6.811 | 1.083 | 0.0 | 0.0 | 5.205 | 0.869 | 3.405 | 7.773 | 449.384 | 15.54 | 0.0 | 0.0 | 0.926 | 0.0 |
| 14 | 0.0 | 1.272 | 3.287 | 0.0 | 1.226 | 3.227 | 0.0 | 3.32 | 47.462 | 29.843 | 367.66 | 17.359 | 2.076 | 0.0 | 14.4 | 1.015 |
| 15 | 9.051 | 2.159 | 38.582 | 0.973 | 5.171 | 3.433 | 23.695 | 2.045 | 86.179 | 65.3 | 220.592 | 13.276 | 1.138 | 1.902 | 18.654 | 0.0 |
| 16 | 13.759 | 75.87 | 11.911 | 0.0 | 8.835 | 50.916 | 11.095 | 1.947 | 38.348 | 216.444 | 37.499 | 9.231 | 0.973 | 8.489 | 4.755 | 2.077 |
| 17 | 13.878 | 22.248 | 19.933 | 5.856 | 32.578 | 158.803 | 86.728 | 73.61 | 9.717 | 28.631 | 13.765 | 13.147 | 0.0 | 3.827 | 6.672 | 2.756 |
| 18 | 9.648 | 6.29 | 37.289 | 2.946 | 37.808 | 38.001 | 75.475 | 62.226 | 27.534 | 18.639 | 66.752 | 14.173 | 14.187 | 17.636 | 20.636 | 42.908 |
| 19 | 30.042 | 10.947 | 41.759 | 6.429 | 18.993 | 15.696 | 33.195 | 12.681 | 55.41 | 59.782 | 66.603 | 18.357 | 8.453 | 57.156 | 42.48 | 14.164 |
| 20 | 24.677 | 19.366 | 64.469 | 4.386 | 15.256 | 48.012 | 67.613 | 12.701 | 20.43 | 52.613 | 104.965 | 6.03 | 6.599 | 16.098 | 26.203 | 2.732 |
| 21 | 17.068 | 3.204 | 45.693 | 0.996 | 2.874 | 26.253 | 90.3 | 16.662 | 56.141 | 65.085 | 124.235 | 17.788 | 1.118 | 2.727 | 19.971 | 2.033 |
| 22 | 14.72 | 21.093 | 38.586 | 2.802 | 10.245 | 27.666 | 39.728 | 19.63 | 33.397 | 80.207 | 133.044 | 33.552 | 6.111 | 6.949 | 15.684 | 8.735 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.845 | -0.821 | 0.452 | -3.01 | -0.219 | 0.539 | 0.95 | -0.884 | 0.112 | 0.42 | 1.12 | -1.386 | -1.905 | -0.486 | 0.012 | -1.336 |
| 02 | -0.576 | -0.521 | 0.217 | -2.281 | -0.36 | -0.049 | 0.721 | -0.534 | 0.399 | 0.833 | 1.288 | -0.204 | -1.644 | -0.923 | -1.185 | -2.535 |
| 03 | -0.763 | -0.551 | 0.604 | -2.655 | -1.039 | 0.161 | 0.647 | -0.215 | -0.139 | 0.617 | 1.371 | -0.57 | -2.216 | -1.245 | -0.075 | -1.356 |
| 04 | -0.773 | -0.854 | -0.334 | -1.628 | -1.424 | 0.247 | 0.392 | -0.115 | 0.164 | 1.146 | 1.242 | -0.195 | -2.096 | -0.993 | -0.115 | -1.153 |
| 05 | -0.188 | -2.173 | -0.014 | -2.592 | -0.103 | 0.282 | 0.758 | -0.149 | 0.81 | 0.624 | 0.681 | -0.779 | -0.993 | -0.918 | -0.044 | -0.695 |
| 06 | 0.183 | -1.418 | 1.015 | -1.923 | -0.557 | -0.378 | 0.74 | -1.047 | 0.604 | 0.032 | 1.057 | -1.161 | -1.698 | -1.298 | 0.31 | -2.598 |
| 07 | -1.108 | -1.871 | 1.059 | -0.871 | -1.129 | -1.967 | 0.37 | -1.13 | 0.235 | -0.495 | 1.899 | -0.611 | -4.387 | -2.471 | -0.436 | -1.961 |
| 08 | -0.333 | -2.166 | 0.052 | -3.085 | -1.32 | -2.589 | -0.588 | -2.408 | 1.002 | -0.108 | 2.059 | -1.614 | -2.182 | -3.044 | 0.161 | -2.412 |
| 09 | -1.673 | -3.016 | 1.253 | -2.492 | -2.567 | -3.173 | 0.004 | -4.387 | -1.539 | -1.894 | 2.308 | -1.579 | -2.481 | -4.387 | -1.152 | -4.387 |
| 10 | -2.96 | -4.387 | -0.722 | -4.387 | -4.387 | -4.387 | -1.53 | -4.387 | -3.063 | -1.262 | 2.676 | -4.387 | -4.387 | -4.387 | -1.285 | -4.387 |
| 11 | -4.387 | -2.96 | -4.387 | -3.063 | -4.387 | -1.262 | -4.387 | -4.387 | -0.808 | 2.673 | -2.027 | -0.802 | -4.387 | -4.387 | -4.387 | -4.387 |
| 12 | -4.387 | -4.387 | -0.808 | -4.387 | -1.194 | -1.573 | 2.659 | -3.166 | -4.387 | -4.387 | -2.027 | -4.387 | -4.387 | -4.387 | -0.728 | -4.387 |
| 13 | -3.006 | -4.387 | -1.465 | -3.053 | -4.387 | -4.387 | -1.717 | -3.211 | -2.106 | -1.339 | 2.67 | -0.671 | -4.387 | -4.387 | -3.166 | -4.387 |
| 14 | -4.387 | -2.932 | -2.137 | -4.387 | -2.96 | -2.154 | -4.387 | -2.128 | 0.429 | -0.03 | 2.47 | -0.562 | -2.537 | -4.387 | -0.745 | -3.1 |
| 15 | -1.194 | -2.504 | 0.224 | -3.131 | -1.723 | -2.098 | -0.257 | -2.55 | 1.022 | 0.746 | 1.959 | -0.824 | -3.016 | -2.61 | -0.492 | -4.387 |
| 16 | -0.789 | 0.895 | -0.929 | -4.387 | -1.217 | 0.499 | -0.998 | -2.591 | 0.218 | 1.94 | 0.196 | -1.175 | -3.131 | -1.255 | -1.801 | -2.537 |
| 17 | -0.781 | -0.319 | -0.427 | -1.607 | 0.057 | 1.631 | 1.029 | 0.865 | -1.126 | -0.071 | -0.789 | -0.833 | -4.387 | -2.0 | -1.484 | -2.294 |
| 18 | -1.133 | -1.54 | 0.19 | -2.235 | 0.204 | 0.209 | 0.89 | 0.698 | -0.109 | -0.493 | 0.768 | -0.76 | -0.759 | -0.547 | -0.393 | 0.329 |
| 19 | -0.023 | -1.011 | 0.302 | -1.519 | -0.474 | -0.661 | 0.075 | -0.869 | 0.583 | 0.658 | 0.766 | -0.508 | -1.259 | 0.614 | 0.319 | -0.761 |
| 20 | -0.217 | -0.455 | 0.733 | -1.876 | -0.689 | 0.441 | 0.781 | -0.867 | -0.403 | 0.532 | 1.219 | -1.58 | -1.495 | -0.636 | -0.158 | -2.301 |
| 21 | -0.579 | -2.16 | 0.392 | -3.114 | -2.257 | -0.156 | 1.069 | -0.603 | 0.596 | 0.743 | 1.387 | -0.539 | -3.029 | -2.303 | -0.425 | -2.555 |
| 22 | -0.724 | -0.372 | 0.224 | -2.279 | -1.075 | -0.105 | 0.253 | -0.442 | 0.081 | 0.951 | 1.455 | 0.086 | -1.567 | -1.446 | -0.662 | -1.228 |