Model info
| Transcription factor | SP4 (GeneCards) | ||||||||
| Model | SP4_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 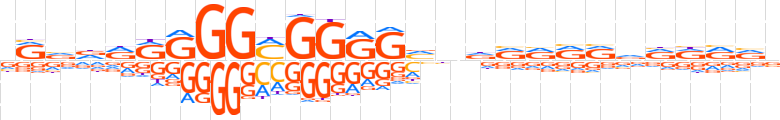 | ||||||||
| LOGO (reverse complement) | 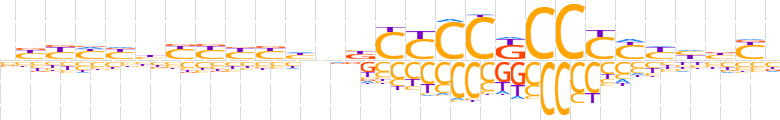 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 27 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | vRvdRRRGGMGGRGvndRRRRvRRRvv | ||||||||
| Best auROC (human) | 0.987 | ||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | 5 | ||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 497 | ||||||||
| TF family | Three-zinc finger Krüppel-related factors {2.3.1} | ||||||||
| TF subfamily | Sp1-like factors {2.3.1.1} | ||||||||
| HGNC | HGNC:11209 | ||||||||
| EntrezGene | GeneID:6671 (SSTAR profile) | ||||||||
| UniProt ID | SP4_HUMAN | ||||||||
| UniProt AC | Q02446 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | SP4 expression | ||||||||
| ReMap ChIP-seq dataset list | SP4 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 16.0 | 10.0 | 70.0 | 4.0 | 29.0 | 12.0 | 55.0 | 14.0 | 30.0 | 22.0 | 179.0 | 6.0 | 3.0 | 0.0 | 40.0 | 2.0 |
| 02 | 16.0 | 13.0 | 43.0 | 6.0 | 11.0 | 9.0 | 18.0 | 6.0 | 95.0 | 46.0 | 167.0 | 36.0 | 7.0 | 0.0 | 17.0 | 2.0 |
| 03 | 54.0 | 4.0 | 60.0 | 11.0 | 14.0 | 5.0 | 27.0 | 22.0 | 135.0 | 20.0 | 73.0 | 17.0 | 16.0 | 5.0 | 23.0 | 6.0 |
| 04 | 73.0 | 6.0 | 128.0 | 12.0 | 14.0 | 4.0 | 13.0 | 3.0 | 84.0 | 16.0 | 66.0 | 17.0 | 8.0 | 1.0 | 42.0 | 5.0 |
| 05 | 30.0 | 1.0 | 133.0 | 15.0 | 3.0 | 1.0 | 20.0 | 3.0 | 38.0 | 6.0 | 157.0 | 48.0 | 8.0 | 0.0 | 22.0 | 7.0 |
| 06 | 14.0 | 0.0 | 61.0 | 4.0 | 3.0 | 0.0 | 5.0 | 0.0 | 101.0 | 7.0 | 221.0 | 3.0 | 1.0 | 1.0 | 71.0 | 0.0 |
| 07 | 2.0 | 0.0 | 117.0 | 0.0 | 2.0 | 0.0 | 5.0 | 1.0 | 1.0 | 1.0 | 355.0 | 1.0 | 0.0 | 0.0 | 7.0 | 0.0 |
| 08 | 2.0 | 0.0 | 3.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 3.0 | 5.0 | 472.0 | 4.0 | 0.0 | 0.0 | 2.0 | 0.0 |
| 09 | 2.0 | 3.0 | 0.0 | 0.0 | 1.0 | 2.0 | 0.0 | 2.0 | 138.0 | 297.0 | 0.0 | 43.0 | 0.0 | 4.0 | 0.0 | 0.0 |
| 10 | 17.0 | 0.0 | 122.0 | 2.0 | 3.0 | 3.0 | 291.0 | 9.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.0 | 0.0 | 41.0 | 1.0 |
| 11 | 1.0 | 0.0 | 19.0 | 3.0 | 0.0 | 0.0 | 3.0 | 0.0 | 10.0 | 9.0 | 407.0 | 28.0 | 0.0 | 0.0 | 11.0 | 1.0 |
| 12 | 4.0 | 0.0 | 6.0 | 1.0 | 2.0 | 0.0 | 7.0 | 0.0 | 125.0 | 1.0 | 300.0 | 14.0 | 1.0 | 1.0 | 30.0 | 0.0 |
| 13 | 13.0 | 0.0 | 116.0 | 3.0 | 1.0 | 0.0 | 1.0 | 0.0 | 60.0 | 6.0 | 269.0 | 8.0 | 2.0 | 2.0 | 11.0 | 0.0 |
| 14 | 25.0 | 34.0 | 13.0 | 4.0 | 3.0 | 1.0 | 2.0 | 2.0 | 104.0 | 213.0 | 46.0 | 34.0 | 1.0 | 7.0 | 3.0 | 0.0 |
| 15 | 41.0 | 23.0 | 48.0 | 21.0 | 21.0 | 96.0 | 40.0 | 98.0 | 24.0 | 17.0 | 6.0 | 17.0 | 3.0 | 14.0 | 7.0 | 16.0 |
| 16 | 17.0 | 9.0 | 53.0 | 10.0 | 40.0 | 14.0 | 54.0 | 42.0 | 38.0 | 8.0 | 48.0 | 7.0 | 34.0 | 9.0 | 80.0 | 29.0 |
| 17 | 29.0 | 10.0 | 84.0 | 6.0 | 10.0 | 10.0 | 15.0 | 5.0 | 42.0 | 26.0 | 155.0 | 12.0 | 23.0 | 9.0 | 44.0 | 12.0 |
| 18 | 18.0 | 7.0 | 74.0 | 5.0 | 26.0 | 6.0 | 20.0 | 3.0 | 86.0 | 32.0 | 155.0 | 25.0 | 11.0 | 1.0 | 22.0 | 1.0 |
| 19 | 30.0 | 17.0 | 89.0 | 5.0 | 7.0 | 5.0 | 31.0 | 3.0 | 67.0 | 36.0 | 156.0 | 12.0 | 12.0 | 1.0 | 18.0 | 3.0 |
| 20 | 27.0 | 6.0 | 77.0 | 6.0 | 15.0 | 14.0 | 26.0 | 4.0 | 63.0 | 34.0 | 186.0 | 11.0 | 3.0 | 3.0 | 14.0 | 3.0 |
| 21 | 28.0 | 8.0 | 60.0 | 12.0 | 19.0 | 6.0 | 19.0 | 13.0 | 96.0 | 62.0 | 118.0 | 27.0 | 9.0 | 1.0 | 10.0 | 4.0 |
| 22 | 43.0 | 5.0 | 99.0 | 5.0 | 17.0 | 8.0 | 39.0 | 13.0 | 55.0 | 29.0 | 105.0 | 18.0 | 8.0 | 4.0 | 38.0 | 6.0 |
| 23 | 20.0 | 14.0 | 82.0 | 7.0 | 6.0 | 6.0 | 21.0 | 13.0 | 52.0 | 25.0 | 177.0 | 27.0 | 6.0 | 3.0 | 21.0 | 12.0 |
| 24 | 22.0 | 6.0 | 53.0 | 3.0 | 10.0 | 7.0 | 27.0 | 4.0 | 122.0 | 34.0 | 134.0 | 11.0 | 8.0 | 7.0 | 44.0 | 0.0 |
| 25 | 41.0 | 16.0 | 102.0 | 3.0 | 10.0 | 8.0 | 29.0 | 7.0 | 66.0 | 41.0 | 129.0 | 22.0 | 1.0 | 3.0 | 11.0 | 3.0 |
| 26 | 27.0 | 26.0 | 54.0 | 11.0 | 24.0 | 10.0 | 30.0 | 4.0 | 34.0 | 56.0 | 154.0 | 27.0 | 6.0 | 11.0 | 14.0 | 4.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.642 | -1.098 | 0.816 | -1.96 | -0.058 | -0.922 | 0.576 | -0.772 | -0.024 | -0.33 | 1.751 | -1.584 | -2.218 | -4.387 | 0.26 | -2.568 |
| 02 | -0.642 | -0.844 | 0.332 | -1.584 | -1.006 | -1.199 | -0.527 | -1.584 | 1.12 | 0.399 | 1.682 | 0.156 | -1.439 | -4.387 | -0.583 | -2.568 |
| 03 | 0.558 | -1.96 | 0.662 | -1.006 | -0.772 | -1.754 | -0.128 | -0.33 | 1.47 | -0.423 | 0.857 | -0.583 | -0.642 | -1.754 | -0.286 | -1.584 |
| 04 | 0.857 | -1.584 | 1.417 | -0.922 | -0.772 | -1.96 | -0.844 | -2.218 | 0.997 | -0.642 | 0.757 | -0.583 | -1.312 | -3.111 | 0.308 | -1.754 |
| 05 | -0.024 | -3.111 | 1.455 | -0.705 | -2.218 | -3.111 | -0.423 | -2.218 | 0.209 | -1.584 | 1.62 | 0.441 | -1.312 | -4.387 | -0.33 | -1.439 |
| 06 | -0.772 | -4.387 | 0.679 | -1.96 | -2.218 | -4.387 | -1.754 | -4.387 | 1.181 | -1.439 | 1.962 | -2.218 | -3.111 | -3.111 | 0.83 | -4.387 |
| 07 | -2.568 | -4.387 | 1.327 | -4.387 | -2.568 | -4.387 | -1.754 | -3.111 | -3.111 | -3.111 | 2.435 | -3.111 | -4.387 | -4.387 | -1.439 | -4.387 |
| 08 | -2.568 | -4.387 | -2.218 | -4.387 | -4.387 | -4.387 | -3.111 | -4.387 | -2.218 | -1.754 | 2.719 | -1.96 | -4.387 | -4.387 | -2.568 | -4.387 |
| 09 | -2.568 | -2.218 | -4.387 | -4.387 | -3.111 | -2.568 | -4.387 | -2.568 | 1.492 | 2.257 | -4.387 | 0.332 | -4.387 | -1.96 | -4.387 | -4.387 |
| 10 | -0.583 | -4.387 | 1.369 | -2.568 | -2.218 | -2.218 | 2.236 | -1.199 | -4.387 | -4.387 | -4.387 | -4.387 | -2.218 | -4.387 | 0.285 | -3.111 |
| 11 | -3.111 | -4.387 | -0.474 | -2.218 | -4.387 | -4.387 | -2.218 | -4.387 | -1.098 | -1.199 | 2.571 | -0.092 | -4.387 | -4.387 | -1.006 | -3.111 |
| 12 | -1.96 | -4.387 | -1.584 | -3.111 | -2.568 | -4.387 | -1.439 | -4.387 | 1.393 | -3.111 | 2.267 | -0.772 | -3.111 | -3.111 | -0.024 | -4.387 |
| 13 | -0.844 | -4.387 | 1.319 | -2.218 | -3.111 | -4.387 | -3.111 | -4.387 | 0.662 | -1.584 | 2.158 | -1.312 | -2.568 | -2.568 | -1.006 | -4.387 |
| 14 | -0.204 | 0.099 | -0.844 | -1.96 | -2.218 | -3.111 | -2.568 | -2.568 | 1.21 | 1.925 | 0.399 | 0.099 | -3.111 | -1.439 | -2.218 | -4.387 |
| 15 | 0.285 | -0.286 | 0.441 | -0.376 | -0.376 | 1.13 | 0.26 | 1.151 | -0.244 | -0.583 | -1.584 | -0.583 | -2.218 | -0.772 | -1.439 | -0.642 |
| 16 | -0.583 | -1.199 | 0.539 | -1.098 | 0.26 | -0.772 | 0.558 | 0.308 | 0.209 | -1.312 | 0.441 | -1.439 | 0.099 | -1.199 | 0.948 | -0.058 |
| 17 | -0.058 | -1.098 | 0.997 | -1.584 | -1.098 | -1.098 | -0.705 | -1.754 | 0.308 | -0.166 | 1.608 | -0.922 | -0.286 | -1.199 | 0.355 | -0.922 |
| 18 | -0.527 | -1.439 | 0.871 | -1.754 | -0.166 | -1.584 | -0.423 | -2.218 | 1.02 | 0.039 | 1.608 | -0.204 | -1.006 | -3.111 | -0.33 | -3.111 |
| 19 | -0.024 | -0.583 | 1.055 | -1.754 | -1.439 | -1.754 | 0.008 | -2.218 | 0.772 | 0.156 | 1.614 | -0.922 | -0.922 | -3.111 | -0.527 | -2.218 |
| 20 | -0.128 | -1.584 | 0.91 | -1.584 | -0.705 | -0.772 | -0.166 | -1.96 | 0.711 | 0.099 | 1.789 | -1.006 | -2.218 | -2.218 | -0.772 | -2.218 |
| 21 | -0.092 | -1.312 | 0.662 | -0.922 | -0.474 | -1.584 | -0.474 | -0.844 | 1.13 | 0.695 | 1.336 | -0.128 | -1.199 | -3.111 | -1.098 | -1.96 |
| 22 | 0.332 | -1.754 | 1.161 | -1.754 | -0.583 | -1.312 | 0.235 | -0.844 | 0.576 | -0.058 | 1.219 | -0.527 | -1.312 | -1.96 | 0.209 | -1.584 |
| 23 | -0.423 | -0.772 | 0.973 | -1.439 | -1.584 | -1.584 | -0.376 | -0.844 | 0.52 | -0.204 | 1.74 | -0.128 | -1.584 | -2.218 | -0.376 | -0.922 |
| 24 | -0.33 | -1.584 | 0.539 | -2.218 | -1.098 | -1.439 | -0.128 | -1.96 | 1.369 | 0.099 | 1.462 | -1.006 | -1.312 | -1.439 | 0.355 | -4.387 |
| 25 | 0.285 | -0.642 | 1.19 | -2.218 | -1.098 | -1.312 | -0.058 | -1.439 | 0.757 | 0.285 | 1.424 | -0.33 | -3.111 | -2.218 | -1.006 | -2.218 |
| 26 | -0.128 | -0.166 | 0.558 | -1.006 | -0.244 | -1.098 | -0.024 | -1.96 | 0.099 | 0.594 | 1.601 | -0.128 | -1.584 | -1.006 | -0.772 | -1.96 |