Model info
| Transcription factor | Sp5 | ||||||||
| Model | SP5_MOUSE.H11DI.0.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 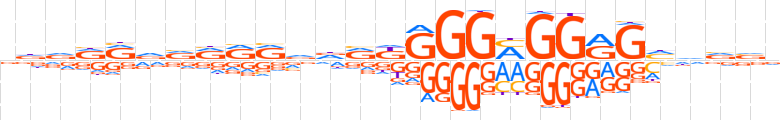 | ||||||||
| LOGO (reverse complement) | 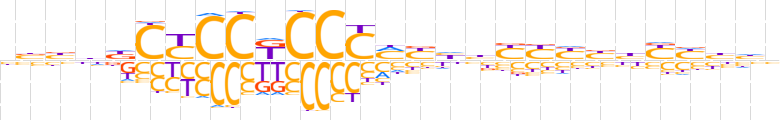 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 27 | ||||||||
| Quality | C | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | dddRRvvRRRvdRKGGGMGGRGhdvvd | ||||||||
| Best auROC (human) | |||||||||
| Best auROC (mouse) | 0.966 | ||||||||
| Peak sets in benchmark (human) | |||||||||
| Peak sets in benchmark (mouse) | 3 | ||||||||
| Aligned words | 511 | ||||||||
| TF family | Three-zinc finger Krüppel-related factors {2.3.1} | ||||||||
| TF subfamily | Sp1-like factors {2.3.1.1} | ||||||||
| MGI | MGI:1927715 | ||||||||
| EntrezGene | GeneID:64406 (SSTAR profile) | ||||||||
| UniProt ID | SP5_MOUSE | ||||||||
| UniProt AC | Q9JHX2 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Sp5 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 24.0 | 6.0 | 68.0 | 11.0 | 21.0 | 14.0 | 20.0 | 20.0 | 51.0 | 19.0 | 116.0 | 46.0 | 11.0 | 9.0 | 34.0 | 26.0 |
| 02 | 20.0 | 9.0 | 75.0 | 3.0 | 11.0 | 13.0 | 14.0 | 10.0 | 98.0 | 18.0 | 95.0 | 27.0 | 11.0 | 13.0 | 61.0 | 18.0 |
| 03 | 18.0 | 9.0 | 104.0 | 9.0 | 14.0 | 11.0 | 8.0 | 20.0 | 44.0 | 25.0 | 156.0 | 20.0 | 8.0 | 8.0 | 33.0 | 9.0 |
| 04 | 15.0 | 8.0 | 60.0 | 1.0 | 13.0 | 11.0 | 9.0 | 20.0 | 44.0 | 24.0 | 223.0 | 10.0 | 4.0 | 10.0 | 36.0 | 8.0 |
| 05 | 24.0 | 14.0 | 36.0 | 2.0 | 24.0 | 8.0 | 14.0 | 7.0 | 136.0 | 37.0 | 126.0 | 29.0 | 6.0 | 5.0 | 23.0 | 5.0 |
| 06 | 36.0 | 16.0 | 127.0 | 11.0 | 27.0 | 4.0 | 21.0 | 12.0 | 47.0 | 38.0 | 95.0 | 19.0 | 5.0 | 3.0 | 30.0 | 5.0 |
| 07 | 19.0 | 14.0 | 71.0 | 11.0 | 14.0 | 9.0 | 31.0 | 7.0 | 58.0 | 31.0 | 165.0 | 19.0 | 8.0 | 2.0 | 29.0 | 8.0 |
| 08 | 10.0 | 5.0 | 79.0 | 5.0 | 23.0 | 5.0 | 24.0 | 4.0 | 86.0 | 26.0 | 170.0 | 14.0 | 3.0 | 3.0 | 35.0 | 4.0 |
| 09 | 23.0 | 6.0 | 91.0 | 2.0 | 10.0 | 6.0 | 15.0 | 8.0 | 47.0 | 48.0 | 195.0 | 18.0 | 2.0 | 2.0 | 23.0 | 0.0 |
| 10 | 15.0 | 5.0 | 55.0 | 7.0 | 19.0 | 20.0 | 10.0 | 13.0 | 117.0 | 60.0 | 119.0 | 28.0 | 3.0 | 1.0 | 19.0 | 5.0 |
| 11 | 59.0 | 5.0 | 60.0 | 30.0 | 59.0 | 6.0 | 8.0 | 13.0 | 113.0 | 24.0 | 35.0 | 31.0 | 16.0 | 11.0 | 20.0 | 6.0 |
| 12 | 56.0 | 20.0 | 163.0 | 8.0 | 6.0 | 17.0 | 19.0 | 4.0 | 31.0 | 13.0 | 57.0 | 22.0 | 11.0 | 11.0 | 56.0 | 2.0 |
| 13 | 7.0 | 1.0 | 82.0 | 14.0 | 17.0 | 1.0 | 11.0 | 32.0 | 56.0 | 7.0 | 158.0 | 74.0 | 5.0 | 0.0 | 21.0 | 10.0 |
| 14 | 15.0 | 0.0 | 70.0 | 0.0 | 6.0 | 0.0 | 2.0 | 1.0 | 75.0 | 4.0 | 193.0 | 0.0 | 15.0 | 0.0 | 113.0 | 2.0 |
| 15 | 2.0 | 0.0 | 107.0 | 2.0 | 0.0 | 0.0 | 3.0 | 1.0 | 5.0 | 5.0 | 364.0 | 4.0 | 0.0 | 0.0 | 1.0 | 2.0 |
| 16 | 1.0 | 1.0 | 2.0 | 3.0 | 1.0 | 0.0 | 3.0 | 1.0 | 10.0 | 0.0 | 460.0 | 5.0 | 0.0 | 0.0 | 6.0 | 3.0 |
| 17 | 9.0 | 3.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 272.0 | 157.0 | 1.0 | 41.0 | 6.0 | 4.0 | 1.0 | 1.0 |
| 18 | 14.0 | 0.0 | 269.0 | 5.0 | 3.0 | 1.0 | 155.0 | 5.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 39.0 | 3.0 |
| 19 | 2.0 | 0.0 | 12.0 | 3.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 2.0 | 438.0 | 24.0 | 0.0 | 3.0 | 10.0 | 0.0 |
| 20 | 1.0 | 0.0 | 1.0 | 1.0 | 3.0 | 1.0 | 2.0 | 0.0 | 224.0 | 1.0 | 226.0 | 9.0 | 2.0 | 1.0 | 24.0 | 0.0 |
| 21 | 2.0 | 2.0 | 220.0 | 6.0 | 0.0 | 0.0 | 1.0 | 2.0 | 27.0 | 10.0 | 207.0 | 9.0 | 0.0 | 3.0 | 6.0 | 1.0 |
| 22 | 8.0 | 16.0 | 3.0 | 2.0 | 3.0 | 6.0 | 2.0 | 4.0 | 119.0 | 229.0 | 38.0 | 48.0 | 4.0 | 6.0 | 0.0 | 8.0 |
| 23 | 47.0 | 13.0 | 62.0 | 12.0 | 115.0 | 30.0 | 30.0 | 82.0 | 26.0 | 7.0 | 5.0 | 5.0 | 16.0 | 12.0 | 16.0 | 18.0 |
| 24 | 38.0 | 32.0 | 119.0 | 15.0 | 20.0 | 15.0 | 17.0 | 10.0 | 42.0 | 9.0 | 52.0 | 10.0 | 17.0 | 21.0 | 66.0 | 13.0 |
| 25 | 17.0 | 12.0 | 77.0 | 11.0 | 26.0 | 23.0 | 13.0 | 15.0 | 37.0 | 48.0 | 143.0 | 26.0 | 6.0 | 7.0 | 28.0 | 7.0 |
| 26 | 13.0 | 15.0 | 48.0 | 10.0 | 30.0 | 15.0 | 20.0 | 25.0 | 78.0 | 30.0 | 118.0 | 35.0 | 10.0 | 13.0 | 32.0 | 4.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.252 | -1.592 | 0.779 | -1.014 | -0.384 | -0.78 | -0.431 | -0.431 | 0.493 | -0.482 | 1.311 | 0.391 | -1.014 | -1.207 | 0.091 | -0.174 |
| 02 | -0.431 | -1.207 | 0.876 | -2.226 | -1.014 | -0.852 | -0.78 | -1.106 | 1.142 | -0.535 | 1.112 | -0.136 | -1.014 | -0.852 | 0.671 | -0.535 |
| 03 | -0.535 | -1.207 | 1.202 | -1.207 | -0.78 | -1.014 | -1.32 | -0.431 | 0.347 | -0.212 | 1.606 | -0.431 | -1.32 | -1.32 | 0.062 | -1.207 |
| 04 | -0.713 | -1.32 | 0.654 | -3.119 | -0.852 | -1.014 | -1.207 | -0.431 | 0.347 | -0.252 | 1.962 | -1.106 | -1.968 | -1.106 | 0.148 | -1.32 |
| 05 | -0.252 | -0.78 | 0.148 | -2.576 | -0.252 | -1.32 | -0.78 | -1.447 | 1.469 | 0.175 | 1.393 | -0.066 | -1.592 | -1.762 | -0.294 | -1.762 |
| 06 | 0.148 | -0.65 | 1.401 | -1.014 | -0.136 | -1.968 | -0.384 | -0.93 | 0.412 | 0.201 | 1.112 | -0.482 | -1.762 | -2.226 | -0.032 | -1.762 |
| 07 | -0.482 | -0.78 | 0.822 | -1.014 | -0.78 | -1.207 | 0.0 | -1.447 | 0.621 | 0.0 | 1.662 | -0.482 | -1.32 | -2.576 | -0.066 | -1.32 |
| 08 | -1.106 | -1.762 | 0.928 | -1.762 | -0.294 | -1.762 | -0.252 | -1.968 | 1.012 | -0.174 | 1.692 | -0.78 | -2.226 | -2.226 | 0.12 | -1.968 |
| 09 | -0.294 | -1.592 | 1.069 | -2.576 | -1.106 | -1.592 | -0.713 | -1.32 | 0.412 | 0.433 | 1.829 | -0.535 | -2.576 | -2.576 | -0.294 | -4.393 |
| 10 | -0.713 | -1.762 | 0.568 | -1.447 | -0.482 | -0.431 | -1.106 | -0.852 | 1.319 | 0.654 | 1.336 | -0.1 | -2.226 | -3.119 | -0.482 | -1.762 |
| 11 | 0.638 | -1.762 | 0.654 | -0.032 | 0.638 | -1.592 | -1.32 | -0.852 | 1.284 | -0.252 | 0.12 | 0.0 | -0.65 | -1.014 | -0.431 | -1.592 |
| 12 | 0.586 | -0.431 | 1.65 | -1.32 | -1.592 | -0.591 | -0.482 | -1.968 | 0.0 | -0.852 | 0.603 | -0.338 | -1.014 | -1.014 | 0.586 | -2.576 |
| 13 | -1.447 | -3.119 | 0.965 | -0.78 | -0.591 | -3.119 | -1.014 | 0.031 | 0.586 | -1.447 | 1.619 | 0.863 | -1.762 | -4.393 | -0.384 | -1.106 |
| 14 | -0.713 | -4.393 | 0.808 | -4.393 | -1.592 | -4.393 | -2.576 | -3.119 | 0.876 | -1.968 | 1.818 | -4.393 | -0.713 | -4.393 | 1.284 | -2.576 |
| 15 | -2.576 | -4.393 | 1.23 | -2.576 | -4.393 | -4.393 | -2.226 | -3.119 | -1.762 | -1.762 | 2.452 | -1.968 | -4.393 | -4.393 | -3.119 | -2.576 |
| 16 | -3.119 | -3.119 | -2.576 | -2.226 | -3.119 | -4.393 | -2.226 | -3.119 | -1.106 | -4.393 | 2.686 | -1.762 | -4.393 | -4.393 | -1.592 | -2.226 |
| 17 | -1.207 | -2.226 | -4.393 | -4.393 | -3.119 | -4.393 | -4.393 | -4.393 | 2.161 | 1.612 | -3.119 | 0.277 | -1.592 | -1.968 | -3.119 | -3.119 |
| 18 | -0.78 | -4.393 | 2.15 | -1.762 | -2.226 | -3.119 | 1.6 | -1.762 | -4.393 | -4.393 | -2.576 | -4.393 | -4.393 | -4.393 | 0.227 | -2.226 |
| 19 | -2.576 | -4.393 | -0.93 | -2.226 | -4.393 | -3.119 | -4.393 | -4.393 | -3.119 | -2.576 | 2.637 | -0.252 | -4.393 | -2.226 | -1.106 | -4.393 |
| 20 | -3.119 | -4.393 | -3.119 | -3.119 | -2.226 | -3.119 | -2.576 | -4.393 | 1.967 | -3.119 | 1.976 | -1.207 | -2.576 | -3.119 | -0.252 | -4.393 |
| 21 | -2.576 | -2.576 | 1.949 | -1.592 | -4.393 | -4.393 | -3.119 | -2.576 | -0.136 | -1.106 | 1.888 | -1.207 | -4.393 | -2.226 | -1.592 | -3.119 |
| 22 | -1.32 | -0.65 | -2.226 | -2.576 | -2.226 | -1.592 | -2.576 | -1.968 | 1.336 | 1.989 | 0.201 | 0.433 | -1.968 | -1.592 | -4.393 | -1.32 |
| 23 | 0.412 | -0.852 | 0.687 | -0.93 | 1.302 | -0.032 | -0.032 | 0.965 | -0.174 | -1.447 | -1.762 | -1.762 | -0.65 | -0.93 | -0.65 | -0.535 |
| 24 | 0.201 | 0.031 | 1.336 | -0.713 | -0.431 | -0.713 | -0.591 | -1.106 | 0.3 | -1.207 | 0.512 | -1.106 | -0.591 | -0.384 | 0.749 | -0.852 |
| 25 | -0.591 | -0.93 | 0.902 | -1.014 | -0.174 | -0.294 | -0.852 | -0.713 | 0.175 | 0.433 | 1.519 | -0.174 | -1.592 | -1.447 | -0.1 | -1.447 |
| 26 | -0.852 | -0.713 | 0.433 | -1.106 | -0.032 | -0.713 | -0.431 | -0.212 | 0.915 | -0.032 | 1.328 | 0.12 | -1.106 | -0.852 | 0.031 | -1.968 |