Model info
| Transcription factor | SPIB (GeneCards) | ||||||||
| Model | SPIB_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 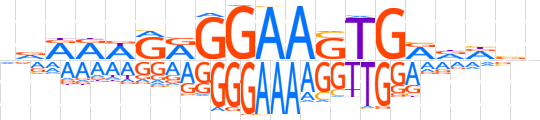 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 19 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | vvRRWRRGGAASTGRdRvn | ||||||||
| Best auROC (human) | 0.999 | ||||||||
| Best auROC (mouse) | 0.961 | ||||||||
| Peak sets in benchmark (human) | 12 | ||||||||
| Peak sets in benchmark (mouse) | 3 | ||||||||
| Aligned words | 413 | ||||||||
| TF family | Ets-related factors {3.5.2} | ||||||||
| TF subfamily | Spi-like factors {3.5.2.5} | ||||||||
| HGNC | HGNC:11242 | ||||||||
| EntrezGene | GeneID:6689 (SSTAR profile) | ||||||||
| UniProt ID | SPIB_HUMAN | ||||||||
| UniProt AC | Q01892 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | SPIB expression | ||||||||
| ReMap ChIP-seq dataset list | SPIB datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 61.0 | 15.0 | 70.0 | 16.0 | 54.0 | 13.0 | 4.0 | 12.0 | 56.0 | 14.0 | 39.0 | 6.0 | 15.0 | 6.0 | 24.0 | 7.0 |
| 02 | 145.0 | 0.0 | 29.0 | 12.0 | 33.0 | 7.0 | 2.0 | 6.0 | 104.0 | 7.0 | 14.0 | 12.0 | 14.0 | 3.0 | 12.0 | 12.0 |
| 03 | 247.0 | 6.0 | 37.0 | 6.0 | 9.0 | 3.0 | 1.0 | 4.0 | 26.0 | 7.0 | 24.0 | 0.0 | 11.0 | 2.0 | 12.0 | 17.0 |
| 04 | 223.0 | 8.0 | 22.0 | 40.0 | 15.0 | 1.0 | 0.0 | 2.0 | 39.0 | 9.0 | 19.0 | 7.0 | 1.0 | 2.0 | 3.0 | 21.0 |
| 05 | 46.0 | 12.0 | 218.0 | 2.0 | 16.0 | 3.0 | 1.0 | 0.0 | 10.0 | 2.0 | 30.0 | 2.0 | 7.0 | 2.0 | 60.0 | 1.0 |
| 06 | 35.0 | 0.0 | 43.0 | 1.0 | 12.0 | 1.0 | 6.0 | 0.0 | 208.0 | 26.0 | 73.0 | 2.0 | 2.0 | 0.0 | 2.0 | 1.0 |
| 07 | 11.0 | 0.0 | 246.0 | 0.0 | 12.0 | 0.0 | 14.0 | 1.0 | 12.0 | 0.0 | 112.0 | 0.0 | 1.0 | 0.0 | 3.0 | 0.0 |
| 08 | 2.0 | 1.0 | 33.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 6.0 | 1.0 | 367.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 |
| 09 | 6.0 | 1.0 | 1.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 392.0 | 8.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 |
| 10 | 392.0 | 1.0 | 3.0 | 4.0 | 7.0 | 0.0 | 1.0 | 1.0 | 3.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 11 | 18.0 | 80.0 | 300.0 | 4.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 2.0 | 2.0 | 0.0 | 0.0 | 1.0 | 4.0 | 0.0 |
| 12 | 3.0 | 1.0 | 0.0 | 14.0 | 10.0 | 1.0 | 0.0 | 73.0 | 7.0 | 7.0 | 2.0 | 290.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| 13 | 2.0 | 0.0 | 19.0 | 0.0 | 1.0 | 0.0 | 9.0 | 0.0 | 0.0 | 0.0 | 3.0 | 0.0 | 2.0 | 7.0 | 367.0 | 2.0 |
| 14 | 3.0 | 0.0 | 2.0 | 0.0 | 2.0 | 1.0 | 2.0 | 2.0 | 263.0 | 30.0 | 89.0 | 16.0 | 0.0 | 0.0 | 2.0 | 0.0 |
| 15 | 175.0 | 15.0 | 70.0 | 8.0 | 12.0 | 3.0 | 4.0 | 12.0 | 16.0 | 16.0 | 41.0 | 22.0 | 1.0 | 5.0 | 9.0 | 3.0 |
| 16 | 160.0 | 10.0 | 18.0 | 16.0 | 15.0 | 6.0 | 1.0 | 17.0 | 69.0 | 15.0 | 25.0 | 15.0 | 8.0 | 10.0 | 18.0 | 9.0 |
| 17 | 40.0 | 92.0 | 101.0 | 19.0 | 13.0 | 14.0 | 4.0 | 10.0 | 9.0 | 21.0 | 22.0 | 10.0 | 5.0 | 15.0 | 24.0 | 13.0 |
| 18 | 18.0 | 13.0 | 27.0 | 9.0 | 28.0 | 56.0 | 9.0 | 49.0 | 34.0 | 42.0 | 32.0 | 43.0 | 11.0 | 9.0 | 14.0 | 18.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.854 | -0.53 | 0.991 | -0.467 | 0.733 | -0.669 | -1.787 | -0.747 | 0.769 | -0.597 | 0.41 | -1.41 | -0.53 | -1.41 | -0.069 | -1.265 |
| 02 | 1.716 | -4.24 | 0.117 | -0.747 | 0.245 | -1.265 | -2.397 | -1.41 | 1.385 | -1.265 | -0.597 | -0.747 | -0.597 | -2.046 | -0.747 | -0.747 |
| 03 | 2.248 | -1.41 | 0.358 | -1.41 | -1.025 | -2.046 | -2.944 | -1.787 | 0.01 | -1.265 | -0.069 | -4.24 | -0.831 | -2.397 | -0.747 | -0.408 |
| 04 | 2.146 | -1.138 | -0.155 | 0.435 | -0.53 | -2.944 | -4.24 | -2.397 | 0.41 | -1.025 | -0.299 | -1.265 | -2.944 | -2.397 | -2.046 | -0.201 |
| 05 | 0.574 | -0.747 | 2.123 | -2.397 | -0.467 | -2.046 | -2.944 | -4.24 | -0.923 | -2.397 | 0.151 | -2.397 | -1.265 | -2.397 | 0.838 | -2.944 |
| 06 | 0.303 | -4.24 | 0.507 | -2.944 | -0.747 | -2.944 | -1.41 | -4.24 | 2.076 | 0.01 | 1.033 | -2.397 | -2.397 | -4.24 | -2.397 | -2.944 |
| 07 | -0.831 | -4.24 | 2.244 | -4.24 | -0.747 | -4.24 | -0.597 | -2.944 | -0.747 | -4.24 | 1.459 | -4.24 | -2.944 | -4.24 | -2.046 | -4.24 |
| 08 | -2.397 | -2.944 | 0.245 | -4.24 | -4.24 | -4.24 | -4.24 | -4.24 | -1.41 | -2.944 | 2.643 | -2.944 | -4.24 | -4.24 | -2.944 | -4.24 |
| 09 | -1.41 | -2.944 | -2.944 | -4.24 | -2.397 | -4.24 | -4.24 | -4.24 | 2.709 | -1.138 | -2.944 | -4.24 | -4.24 | -4.24 | -2.944 | -4.24 |
| 10 | 2.709 | -2.944 | -2.046 | -1.787 | -1.265 | -4.24 | -2.944 | -2.944 | -2.046 | -4.24 | -4.24 | -4.24 | -4.24 | -4.24 | -4.24 | -4.24 |
| 11 | -0.352 | 1.124 | 2.442 | -1.787 | -4.24 | -2.944 | -4.24 | -4.24 | -4.24 | -2.397 | -2.397 | -4.24 | -4.24 | -2.944 | -1.787 | -4.24 |
| 12 | -2.046 | -2.944 | -4.24 | -0.597 | -0.923 | -2.944 | -4.24 | 1.033 | -1.265 | -1.265 | -2.397 | 2.408 | -2.944 | -2.944 | -2.944 | -2.944 |
| 13 | -2.397 | -4.24 | -0.299 | -4.24 | -2.944 | -4.24 | -1.025 | -4.24 | -4.24 | -4.24 | -2.046 | -4.24 | -2.397 | -1.265 | 2.643 | -2.397 |
| 14 | -2.046 | -4.24 | -2.397 | -4.24 | -2.397 | -2.944 | -2.397 | -2.397 | 2.311 | 0.151 | 1.23 | -0.467 | -4.24 | -4.24 | -2.397 | -4.24 |
| 15 | 1.904 | -0.53 | 0.991 | -1.138 | -0.747 | -2.046 | -1.787 | -0.747 | -0.467 | -0.467 | 0.46 | -0.155 | -2.944 | -1.581 | -1.025 | -2.046 |
| 16 | 1.815 | -0.923 | -0.352 | -0.467 | -0.53 | -1.41 | -2.944 | -0.408 | 0.977 | -0.53 | -0.029 | -0.53 | -1.138 | -0.923 | -0.352 | -1.025 |
| 17 | 0.435 | 1.263 | 1.356 | -0.299 | -0.669 | -0.597 | -1.787 | -0.923 | -1.025 | -0.201 | -0.155 | -0.923 | -1.581 | -0.53 | -0.069 | -0.669 |
| 18 | -0.352 | -0.669 | 0.047 | -1.025 | 0.083 | 0.769 | -1.025 | 0.637 | 0.274 | 0.484 | 0.214 | 0.507 | -0.831 | -1.025 | -0.597 | -0.352 |