Model info
| Transcription factor | Srf | ||||||||
| Model | SRF_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 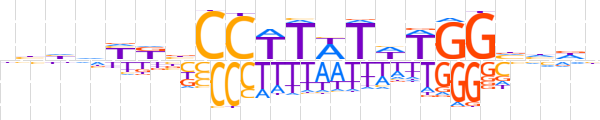 | ||||||||
| LOGO (reverse complement) | 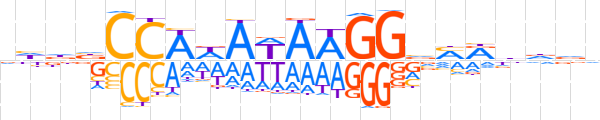 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 21 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | hhhdYKbCCTTWTWTGGvhdb | ||||||||
| Best auROC (human) | 0.951 | ||||||||
| Best auROC (mouse) | 0.988 | ||||||||
| Peak sets in benchmark (human) | 46 | ||||||||
| Peak sets in benchmark (mouse) | 37 | ||||||||
| Aligned words | 322 | ||||||||
| TF family | Responders to external signals (SRF/RLM1) {5.1.2} | ||||||||
| TF subfamily | SRF {5.1.2.0.1} | ||||||||
| MGI | MGI:106658 | ||||||||
| EntrezGene | GeneID:20807 (SSTAR profile) | ||||||||
| UniProt ID | SRF_MOUSE | ||||||||
| UniProt AC | Q9JM73 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Srf expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 19.0 | 20.0 | 17.0 | 13.0 | 13.0 | 35.0 | 8.0 | 17.0 | 10.0 | 18.0 | 10.0 | 11.0 | 10.0 | 71.0 | 14.0 | 29.0 |
| 02 | 7.0 | 13.0 | 12.0 | 20.0 | 28.0 | 22.0 | 7.0 | 87.0 | 11.0 | 8.0 | 13.0 | 17.0 | 10.0 | 21.0 | 8.0 | 31.0 |
| 03 | 18.0 | 14.0 | 15.0 | 9.0 | 29.0 | 14.0 | 10.0 | 11.0 | 18.0 | 9.0 | 8.0 | 5.0 | 65.0 | 15.0 | 41.0 | 34.0 |
| 04 | 19.0 | 16.0 | 10.0 | 85.0 | 12.0 | 6.0 | 3.0 | 31.0 | 8.0 | 18.0 | 5.0 | 43.0 | 3.0 | 6.0 | 4.0 | 46.0 |
| 05 | 5.0 | 5.0 | 22.0 | 10.0 | 5.0 | 2.0 | 6.0 | 33.0 | 4.0 | 5.0 | 1.0 | 12.0 | 9.0 | 9.0 | 63.0 | 124.0 |
| 06 | 2.0 | 6.0 | 11.0 | 4.0 | 0.0 | 8.0 | 1.0 | 12.0 | 13.0 | 47.0 | 13.0 | 19.0 | 12.0 | 58.0 | 54.0 | 55.0 |
| 07 | 2.0 | 24.0 | 0.0 | 1.0 | 1.0 | 116.0 | 0.0 | 2.0 | 1.0 | 76.0 | 1.0 | 1.0 | 1.0 | 84.0 | 1.0 | 4.0 |
| 08 | 0.0 | 2.0 | 2.0 | 1.0 | 1.0 | 283.0 | 1.0 | 15.0 | 1.0 | 1.0 | 0.0 | 0.0 | 1.0 | 6.0 | 0.0 | 1.0 |
| 09 | 0.0 | 0.0 | 0.0 | 3.0 | 74.0 | 2.0 | 1.0 | 215.0 | 0.0 | 1.0 | 0.0 | 2.0 | 7.0 | 3.0 | 1.0 | 6.0 |
| 10 | 5.0 | 3.0 | 1.0 | 72.0 | 0.0 | 3.0 | 0.0 | 3.0 | 0.0 | 0.0 | 0.0 | 2.0 | 15.0 | 8.0 | 1.0 | 202.0 |
| 11 | 17.0 | 2.0 | 0.0 | 1.0 | 1.0 | 2.0 | 1.0 | 10.0 | 0.0 | 0.0 | 2.0 | 0.0 | 188.0 | 13.0 | 4.0 | 74.0 |
| 12 | 15.0 | 2.0 | 2.0 | 187.0 | 4.0 | 2.0 | 0.0 | 11.0 | 0.0 | 0.0 | 0.0 | 7.0 | 4.0 | 1.0 | 6.0 | 74.0 |
| 13 | 17.0 | 1.0 | 3.0 | 2.0 | 0.0 | 1.0 | 0.0 | 4.0 | 1.0 | 2.0 | 4.0 | 1.0 | 99.0 | 21.0 | 8.0 | 151.0 |
| 14 | 25.0 | 1.0 | 10.0 | 81.0 | 5.0 | 0.0 | 0.0 | 20.0 | 1.0 | 0.0 | 0.0 | 14.0 | 16.0 | 0.0 | 12.0 | 130.0 |
| 15 | 4.0 | 0.0 | 43.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 2.0 | 0.0 | 19.0 | 1.0 | 25.0 | 1.0 | 215.0 | 4.0 |
| 16 | 3.0 | 1.0 | 26.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 269.0 | 8.0 | 1.0 | 0.0 | 0.0 | 4.0 |
| 17 | 0.0 | 4.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 50.0 | 156.0 | 51.0 | 39.0 | 3.0 | 6.0 | 0.0 | 4.0 |
| 18 | 21.0 | 15.0 | 7.0 | 11.0 | 52.0 | 64.0 | 7.0 | 43.0 | 14.0 | 24.0 | 3.0 | 11.0 | 10.0 | 17.0 | 8.0 | 8.0 |
| 19 | 35.0 | 5.0 | 40.0 | 17.0 | 90.0 | 12.0 | 3.0 | 15.0 | 9.0 | 5.0 | 8.0 | 3.0 | 25.0 | 6.0 | 12.0 | 30.0 |
| 20 | 22.0 | 59.0 | 23.0 | 55.0 | 1.0 | 10.0 | 5.0 | 12.0 | 11.0 | 22.0 | 10.0 | 20.0 | 4.0 | 11.0 | 24.0 | 26.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.035 | 0.015 | -0.144 | -0.406 | -0.406 | 0.567 | -0.875 | -0.144 | -0.66 | -0.088 | -0.66 | -0.568 | -0.66 | 1.27 | -0.334 | 0.382 |
| 02 | -1.002 | -0.406 | -0.484 | 0.015 | 0.347 | 0.109 | -1.002 | 1.472 | -0.568 | -0.875 | -0.406 | -0.144 | -0.66 | 0.063 | -0.875 | 0.447 |
| 03 | -0.088 | -0.334 | -0.266 | -0.762 | 0.382 | -0.334 | -0.66 | -0.568 | -0.088 | -0.762 | -0.875 | -1.319 | 1.182 | -0.266 | 0.724 | 0.539 |
| 04 | -0.035 | -0.203 | -0.66 | 1.449 | -0.484 | -1.148 | -1.786 | 0.447 | -0.875 | -0.088 | -1.319 | 0.771 | -1.786 | -1.148 | -1.526 | 0.838 |
| 05 | -1.319 | -1.319 | 0.109 | -0.66 | -1.319 | -2.14 | -1.148 | 0.509 | -1.526 | -1.319 | -2.691 | -0.484 | -0.762 | -0.762 | 1.151 | 1.825 |
| 06 | -2.14 | -1.148 | -0.568 | -1.526 | -4.021 | -0.875 | -2.691 | -0.484 | -0.406 | 0.86 | -0.406 | -0.035 | -0.484 | 1.069 | 0.998 | 1.016 |
| 07 | -2.14 | 0.195 | -4.021 | -2.691 | -2.691 | 1.759 | -4.021 | -2.14 | -2.691 | 1.337 | -2.691 | -2.691 | -2.691 | 1.437 | -2.691 | -1.526 |
| 08 | -4.021 | -2.14 | -2.14 | -2.691 | -2.691 | 2.649 | -2.691 | -0.266 | -2.691 | -2.691 | -4.021 | -4.021 | -2.691 | -1.148 | -4.021 | -2.691 |
| 09 | -4.021 | -4.021 | -4.021 | -1.786 | 1.311 | -2.14 | -2.691 | 2.374 | -4.021 | -2.691 | -4.021 | -2.14 | -1.002 | -1.786 | -2.691 | -1.148 |
| 10 | -1.319 | -1.786 | -2.691 | 1.284 | -4.021 | -1.786 | -4.021 | -1.786 | -4.021 | -4.021 | -4.021 | -2.14 | -0.266 | -0.875 | -2.691 | 2.312 |
| 11 | -0.144 | -2.14 | -4.021 | -2.691 | -2.691 | -2.14 | -2.691 | -0.66 | -4.021 | -4.021 | -2.14 | -4.021 | 2.24 | -0.406 | -1.526 | 1.311 |
| 12 | -0.266 | -2.14 | -2.14 | 2.235 | -1.526 | -2.14 | -4.021 | -0.568 | -4.021 | -4.021 | -4.021 | -1.002 | -1.526 | -2.691 | -1.148 | 1.311 |
| 13 | -0.144 | -2.691 | -1.786 | -2.14 | -4.021 | -2.691 | -4.021 | -1.526 | -2.691 | -2.14 | -1.526 | -2.691 | 1.601 | 0.063 | -0.875 | 2.022 |
| 14 | 0.235 | -2.691 | -0.66 | 1.401 | -1.319 | -4.021 | -4.021 | 0.015 | -2.691 | -4.021 | -4.021 | -0.334 | -0.203 | -4.021 | -0.484 | 1.872 |
| 15 | -1.526 | -4.021 | 0.771 | -4.021 | -4.021 | -4.021 | -2.691 | -4.021 | -2.14 | -4.021 | -0.035 | -2.691 | 0.235 | -2.691 | 2.374 | -1.526 |
| 16 | -1.786 | -2.691 | 0.274 | -2.691 | -4.021 | -4.021 | -2.691 | -4.021 | -2.691 | -4.021 | 2.598 | -0.875 | -2.691 | -4.021 | -4.021 | -1.526 |
| 17 | -4.021 | -1.526 | -2.691 | -4.021 | -2.691 | -4.021 | -4.021 | -4.021 | 0.921 | 2.054 | 0.941 | 0.675 | -1.786 | -1.148 | -4.021 | -1.526 |
| 18 | 0.063 | -0.266 | -1.002 | -0.568 | 0.96 | 1.166 | -1.002 | 0.771 | -0.334 | 0.195 | -1.786 | -0.568 | -0.66 | -0.144 | -0.875 | -0.875 |
| 19 | 0.567 | -1.319 | 0.7 | -0.144 | 1.506 | -0.484 | -1.786 | -0.266 | -0.762 | -1.319 | -0.875 | -1.786 | 0.235 | -1.148 | -0.484 | 0.415 |
| 20 | 0.109 | 1.086 | 0.153 | 1.016 | -2.691 | -0.66 | -1.319 | -0.484 | -0.568 | 0.109 | -0.66 | 0.015 | -1.526 | -0.568 | 0.195 | 0.274 |