Model info
| Transcription factor | STAT3 (GeneCards) | ||||||||
| Model | STAT3_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 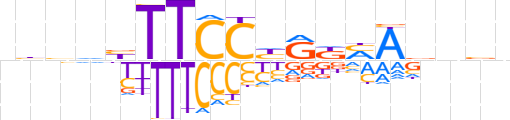 | ||||||||
| LOGO (reverse complement) | 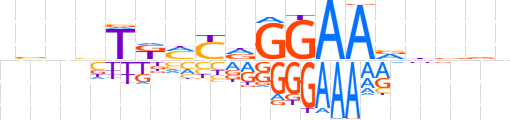 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv10 | ||||||||
| Model length | 18 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nbbvbTTCChRKMAvnbn | ||||||||
| Best auROC (human) | 0.956 | ||||||||
| Best auROC (mouse) | 0.952 | ||||||||
| Peak sets in benchmark (human) | 36 | ||||||||
| Peak sets in benchmark (mouse) | 82 | ||||||||
| Aligned words | 436 | ||||||||
| TF family | STAT factors {6.2.1} | ||||||||
| TF subfamily | STAT3 {6.2.1.0.3} | ||||||||
| HGNC | HGNC:11364 | ||||||||
| EntrezGene | GeneID:6774 (SSTAR profile) | ||||||||
| UniProt ID | STAT3_HUMAN | ||||||||
| UniProt AC | P40763 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | STAT3 expression | ||||||||
| ReMap ChIP-seq dataset list | STAT3 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 6.027 | 31.091 | 31.987 | 16.977 | 21.924 | 37.09 | 7.078 | 49.745 | 9.124 | 33.821 | 32.891 | 49.06 | 11.827 | 43.198 | 26.4 | 27.757 |
| 02 | 3.875 | 19.333 | 18.842 | 6.853 | 32.178 | 55.483 | 11.085 | 46.455 | 14.633 | 30.096 | 25.238 | 28.389 | 7.087 | 69.703 | 34.597 | 32.152 |
| 03 | 15.875 | 10.655 | 23.363 | 7.881 | 93.977 | 32.705 | 16.177 | 31.757 | 33.157 | 15.376 | 32.536 | 8.694 | 19.921 | 26.083 | 58.775 | 9.072 |
| 04 | 3.826 | 47.596 | 29.954 | 81.553 | 2.856 | 33.888 | 2.921 | 45.153 | 12.199 | 39.105 | 19.165 | 60.381 | 4.881 | 17.574 | 13.186 | 21.764 |
| 05 | 0.0 | 0.926 | 0.0 | 22.836 | 0.0 | 5.184 | 0.0 | 132.979 | 1.015 | 1.004 | 0.0 | 63.207 | 0.0 | 0.991 | 0.0 | 207.859 |
| 06 | 0.0 | 0.0 | 0.0 | 1.015 | 0.0 | 0.0 | 0.0 | 8.105 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.059 | 424.821 |
| 07 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.059 | 0.0 | 0.0 | 45.991 | 385.869 | 0.0 | 2.081 |
| 08 | 0.0 | 45.991 | 0.0 | 0.0 | 0.0 | 325.65 | 0.976 | 61.301 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.081 | 0.0 | 0.0 |
| 09 | 0.0 | 0.0 | 0.0 | 0.0 | 48.26 | 155.005 | 15.173 | 155.285 | 0.976 | 0.0 | 0.0 | 0.0 | 5.006 | 37.467 | 2.994 | 15.834 |
| 10 | 2.959 | 0.0 | 51.283 | 0.0 | 99.379 | 9.938 | 72.058 | 11.096 | 6.135 | 1.07 | 10.961 | 0.0 | 8.911 | 0.0 | 161.256 | 0.952 |
| 11 | 7.115 | 5.777 | 89.336 | 15.155 | 1.929 | 1.07 | 3.033 | 4.976 | 26.154 | 20.995 | 141.492 | 106.918 | 0.0 | 1.022 | 6.96 | 4.068 |
| 12 | 22.029 | 11.093 | 0.0 | 2.076 | 14.316 | 9.634 | 0.0 | 4.914 | 128.327 | 91.911 | 3.964 | 16.619 | 62.166 | 44.865 | 4.883 | 19.203 |
| 13 | 204.036 | 8.068 | 7.851 | 6.883 | 130.697 | 8.681 | 1.963 | 16.162 | 5.908 | 1.051 | 0.95 | 0.939 | 30.612 | 4.989 | 2.117 | 5.094 |
| 14 | 66.539 | 93.122 | 160.883 | 50.709 | 4.145 | 11.898 | 1.018 | 5.727 | 0.933 | 9.935 | 0.971 | 1.042 | 6.015 | 3.103 | 14.017 | 5.943 |
| 15 | 16.99 | 12.042 | 27.437 | 21.164 | 18.786 | 35.874 | 6.791 | 56.607 | 46.971 | 46.115 | 21.776 | 62.026 | 7.814 | 22.689 | 12.949 | 19.969 |
| 16 | 15.816 | 25.044 | 40.962 | 8.739 | 20.908 | 43.705 | 16.189 | 35.918 | 6.856 | 23.93 | 24.918 | 13.248 | 11.869 | 48.603 | 74.805 | 24.489 |
| 17 | 12.648 | 13.944 | 21.991 | 6.866 | 58.581 | 40.252 | 11.859 | 30.59 | 54.304 | 24.086 | 49.435 | 29.05 | 11.073 | 22.618 | 33.699 | 15.006 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -1.462 | 0.13 | 0.158 | -0.465 | -0.214 | 0.305 | -1.31 | 0.596 | -1.067 | 0.213 | 0.186 | 0.582 | -0.817 | 0.456 | -0.031 | 0.018 |
| 02 | -1.871 | -0.338 | -0.363 | -1.34 | 0.164 | 0.704 | -0.88 | 0.528 | -0.61 | 0.098 | -0.076 | 0.04 | -1.308 | 0.931 | 0.236 | 0.163 |
| 03 | -0.531 | -0.918 | -0.152 | -1.207 | 1.228 | 0.18 | -0.512 | 0.151 | 0.194 | -0.562 | 0.175 | -1.113 | -0.308 | -0.043 | 0.761 | -1.073 |
| 04 | -1.882 | 0.552 | 0.093 | 1.087 | -2.145 | 0.215 | -2.125 | 0.5 | -0.787 | 0.357 | -0.346 | 0.788 | -1.659 | -0.431 | -0.711 | -0.221 |
| 05 | -4.287 | -3.052 | -4.287 | -0.174 | -4.287 | -1.603 | -4.287 | 1.574 | -2.986 | -2.994 | -4.287 | 0.834 | -4.287 | -3.003 | -4.287 | 2.02 |
| 06 | -4.287 | -4.287 | -4.287 | -2.986 | -4.287 | -4.287 | -4.287 | -1.181 | -4.287 | -4.287 | -4.287 | -4.287 | -4.287 | -4.287 | -2.427 | 2.734 |
| 07 | -4.287 | -4.287 | -4.287 | -4.287 | -4.287 | -4.287 | -4.287 | -4.287 | -4.287 | -2.427 | -4.287 | -4.287 | 0.518 | 2.638 | -4.287 | -2.418 |
| 08 | -4.287 | 0.518 | -4.287 | -4.287 | -4.287 | 2.468 | -3.014 | 0.803 | -4.287 | -4.287 | -4.287 | -4.287 | -4.287 | -2.418 | -4.287 | -4.287 |
| 09 | -4.287 | -4.287 | -4.287 | -4.287 | 0.566 | 1.727 | -0.575 | 1.729 | -3.014 | -4.287 | -4.287 | -4.287 | -1.635 | 0.315 | -2.103 | -0.533 |
| 10 | -2.113 | -4.287 | 0.626 | -4.287 | 1.284 | -0.985 | 0.964 | -0.879 | -1.445 | -2.947 | -0.89 | -4.287 | -1.09 | -4.287 | 1.766 | -3.032 |
| 11 | -1.305 | -1.501 | 1.178 | -0.576 | -2.482 | -2.947 | -2.091 | -1.641 | -0.04 | -0.257 | 1.636 | 1.357 | -4.287 | -2.981 | -1.326 | -1.827 |
| 12 | -0.209 | -0.879 | -4.287 | -2.421 | -0.631 | -1.015 | -4.287 | -1.652 | 1.539 | 1.206 | -1.85 | -0.486 | 0.817 | 0.493 | -1.658 | -0.344 |
| 13 | 2.001 | -1.185 | -1.211 | -1.336 | 1.557 | -1.115 | -2.467 | -0.513 | -1.48 | -2.961 | -3.034 | -3.042 | 0.115 | -1.638 | -2.404 | -1.619 |
| 14 | 0.885 | 1.219 | 1.764 | 0.615 | -1.809 | -0.811 | -2.984 | -1.51 | -3.046 | -0.985 | -3.018 | -2.967 | -1.463 | -2.071 | -0.652 | -1.475 |
| 15 | -0.464 | -0.799 | 0.007 | -0.249 | -0.366 | 0.272 | -1.349 | 0.724 | 0.539 | 0.52 | -0.221 | 0.815 | -1.216 | -0.18 | -0.729 | -0.306 |
| 16 | -0.534 | -0.083 | 0.403 | -1.109 | -0.261 | 0.467 | -0.511 | 0.273 | -1.34 | -0.128 | -0.088 | -0.707 | -0.813 | 0.573 | 1.001 | -0.105 |
| 17 | -0.752 | -0.657 | -0.211 | -1.338 | 0.758 | 0.386 | -0.814 | 0.114 | 0.683 | -0.122 | 0.589 | 0.063 | -0.881 | -0.184 | 0.21 | -0.585 |