Model info
| Transcription factor | TAL1 (GeneCards) | ||||||||
| Model | TAL1_HUMAN.H11MO.0.A | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO | 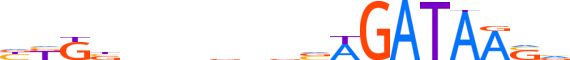 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 19 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | bKKbnnnnnvvWGATAASv | ||||||||
| Best auROC (human) | 0.978 | ||||||||
| Best auROC (mouse) | 0.968 | ||||||||
| Peak sets in benchmark (human) | 68 | ||||||||
| Peak sets in benchmark (mouse) | 76 | ||||||||
| Aligned words | 500 | ||||||||
| TF family | Tal-related factors {1.2.3} | ||||||||
| TF subfamily | Tal / HEN-like factors {1.2.3.1} | ||||||||
| HGNC | HGNC:11556 | ||||||||
| EntrezGene | GeneID:6886 (SSTAR profile) | ||||||||
| UniProt ID | TAL1_HUMAN | ||||||||
| UniProt AC | P17542 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | TAL1 expression | ||||||||
| ReMap ChIP-seq dataset list | TAL1 datasets | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 51.0 | 286.0 | 80.0 | 83.0 |
| 02 | 35.0 | 74.0 | 76.0 | 315.0 |
| 03 | 17.0 | 18.0 | 360.0 | 105.0 |
| 04 | 29.0 | 135.0 | 242.0 | 94.0 |
| 05 | 93.0 | 113.0 | 162.0 | 132.0 |
| 06 | 115.0 | 113.0 | 182.0 | 90.0 |
| 07 | 107.0 | 107.0 | 193.0 | 93.0 |
| 08 | 96.0 | 105.0 | 203.0 | 96.0 |
| 09 | 77.0 | 154.0 | 159.0 | 110.0 |
| 10 | 145.0 | 85.0 | 189.0 | 81.0 |
| 11 | 59.0 | 237.0 | 164.0 | 40.0 |
| 12 | 320.0 | 16.0 | 4.0 | 160.0 |
| 13 | 1.0 | 1.0 | 498.0 | 0.0 |
| 14 | 500.0 | 0.0 | 0.0 | 0.0 |
| 15 | 2.0 | 0.0 | 1.0 | 497.0 |
| 16 | 475.0 | 1.0 | 6.0 | 18.0 |
| 17 | 411.0 | 8.0 | 74.0 | 7.0 |
| 18 | 72.0 | 82.0 | 317.0 | 29.0 |
| 19 | 145.0 | 109.0 | 207.0 | 39.0 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.879 | 0.821 | -0.439 | -0.403 |
| 02 | -1.242 | -0.516 | -0.49 | 0.917 |
| 03 | -1.92 | -1.868 | 1.05 | -0.172 |
| 04 | -1.421 | 0.076 | 0.655 | -0.281 |
| 05 | -0.291 | -0.1 | 0.256 | 0.054 |
| 06 | -0.082 | -0.1 | 0.372 | -0.324 |
| 07 | -0.153 | -0.153 | 0.43 | -0.291 |
| 08 | -0.26 | -0.172 | 0.48 | -0.26 |
| 09 | -0.477 | 0.206 | 0.238 | -0.126 |
| 10 | 0.147 | -0.38 | 0.409 | -0.427 |
| 11 | -0.737 | 0.634 | 0.269 | -1.114 |
| 12 | 0.932 | -1.975 | -3.126 | 0.244 |
| 13 | -3.903 | -3.903 | 1.373 | -4.4 |
| 14 | 1.377 | -4.4 | -4.4 | -4.4 |
| 15 | -3.573 | -4.4 | -3.903 | 1.371 |
| 16 | 1.326 | -3.903 | -2.819 | -1.868 |
| 17 | 1.182 | -2.584 | -0.516 | -2.694 |
| 18 | -0.543 | -0.415 | 0.923 | -1.421 |
| 19 | 0.147 | -0.135 | 0.5 | -1.138 |