Model info
| Transcription factor | Tead1 | ||||||||
| Model | TEAD1_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 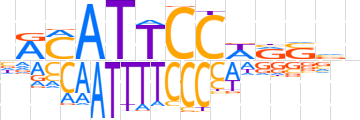 | ||||||||
| LOGO (reverse complement) | 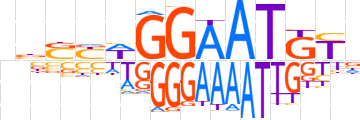 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 13 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nRMATTCCWRSvn | ||||||||
| Best auROC (human) | 0.961 | ||||||||
| Best auROC (mouse) | 0.952 | ||||||||
| Peak sets in benchmark (human) | 14 | ||||||||
| Peak sets in benchmark (mouse) | 5 | ||||||||
| Aligned words | 439 | ||||||||
| TF family | TEF-1-related factors {3.6.1} | ||||||||
| TF subfamily | TEF-1 (TEAD-1, TCF-13) {3.6.1.0.1} | ||||||||
| MGI | MGI:101876 | ||||||||
| EntrezGene | GeneID:21676 (SSTAR profile) | ||||||||
| UniProt ID | TEAD1_MOUSE | ||||||||
| UniProt AC | P30051 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Tead1 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 57.0 | 0.0 | 38.0 | 0.0 | 126.0 | 1.0 | 25.0 | 5.0 | 67.0 | 3.0 | 22.0 | 1.0 | 39.0 | 2.0 | 51.0 | 1.0 |
| 02 | 74.0 | 207.0 | 8.0 | 0.0 | 1.0 | 5.0 | 0.0 | 0.0 | 27.0 | 100.0 | 8.0 | 1.0 | 0.0 | 7.0 | 0.0 | 0.0 |
| 03 | 99.0 | 2.0 | 1.0 | 0.0 | 315.0 | 3.0 | 1.0 | 0.0 | 16.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 |
| 04 | 0.0 | 1.0 | 1.0 | 429.0 | 0.0 | 0.0 | 0.0 | 5.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 05 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 49.0 | 1.0 | 9.0 | 377.0 |
| 06 | 0.0 | 45.0 | 5.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 9.0 | 0.0 | 0.0 | 0.0 | 369.0 | 5.0 | 3.0 |
| 07 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 392.0 | 1.0 | 31.0 | 0.0 | 10.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 2.0 |
| 08 | 0.0 | 0.0 | 0.0 | 1.0 | 235.0 | 17.0 | 26.0 | 125.0 | 0.0 | 0.0 | 1.0 | 0.0 | 2.0 | 1.0 | 4.0 | 26.0 |
| 09 | 41.0 | 14.0 | 174.0 | 8.0 | 7.0 | 6.0 | 1.0 | 4.0 | 2.0 | 12.0 | 11.0 | 6.0 | 14.0 | 22.0 | 99.0 | 17.0 |
| 10 | 0.0 | 10.0 | 52.0 | 2.0 | 11.0 | 15.0 | 25.0 | 3.0 | 15.0 | 90.0 | 165.0 | 15.0 | 1.0 | 5.0 | 27.0 | 2.0 |
| 11 | 6.0 | 7.0 | 12.0 | 2.0 | 45.0 | 47.0 | 9.0 | 19.0 | 52.0 | 86.0 | 120.0 | 11.0 | 1.0 | 1.0 | 17.0 | 3.0 |
| 12 | 39.0 | 20.0 | 33.0 | 12.0 | 67.0 | 25.0 | 18.0 | 31.0 | 41.0 | 48.0 | 39.0 | 30.0 | 3.0 | 5.0 | 22.0 | 5.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.726 | -4.291 | 0.324 | -4.291 | 1.516 | -3.001 | -0.089 | -1.641 | 0.887 | -2.106 | -0.215 | -3.001 | 0.35 | -2.456 | 0.616 | -3.001 |
| 02 | 0.986 | 2.011 | -1.198 | -4.291 | -3.001 | -1.641 | -4.291 | -4.291 | -0.014 | 1.286 | -1.198 | -3.001 | -4.291 | -1.325 | -4.291 | -4.291 |
| 03 | 1.276 | -2.456 | -3.001 | -4.291 | 2.43 | -2.106 | -3.001 | -4.291 | -0.527 | -4.291 | -4.291 | -4.291 | -3.001 | -4.291 | -4.291 | -4.291 |
| 04 | -4.291 | -3.001 | -3.001 | 2.739 | -4.291 | -4.291 | -4.291 | -1.641 | -4.291 | -4.291 | -4.291 | -2.456 | -4.291 | -4.291 | -4.291 | -4.291 |
| 05 | -4.291 | -4.291 | -4.291 | -4.291 | -3.001 | -4.291 | -4.291 | -4.291 | -4.291 | -3.001 | -4.291 | -4.291 | 0.576 | -3.001 | -1.085 | 2.61 |
| 06 | -4.291 | 0.492 | -1.641 | -4.291 | -4.291 | -2.456 | -4.291 | -4.291 | -4.291 | -1.085 | -4.291 | -4.291 | -4.291 | 2.588 | -1.641 | -2.106 |
| 07 | -4.291 | -4.291 | -4.291 | -4.291 | -3.001 | 2.649 | -3.001 | 0.123 | -4.291 | -0.984 | -4.291 | -4.291 | -4.291 | -3.001 | -4.291 | -2.456 |
| 08 | -4.291 | -4.291 | -4.291 | -3.001 | 2.138 | -0.468 | -0.051 | 1.508 | -4.291 | -4.291 | -3.001 | -4.291 | -2.456 | -3.001 | -1.846 | -0.051 |
| 09 | 0.399 | -0.658 | 1.838 | -1.198 | -1.325 | -1.47 | -3.001 | -1.846 | -2.456 | -0.807 | -0.892 | -1.47 | -0.658 | -0.215 | 1.276 | -0.468 |
| 10 | -4.291 | -0.984 | 0.635 | -2.456 | -0.892 | -0.59 | -0.089 | -2.106 | -0.59 | 1.181 | 1.785 | -0.59 | -3.001 | -1.641 | -0.014 | -2.456 |
| 11 | -1.47 | -1.325 | -0.807 | -2.456 | 0.492 | 0.535 | -1.085 | -0.359 | 0.635 | 1.135 | 1.467 | -0.892 | -3.001 | -3.001 | -0.468 | -2.106 |
| 12 | 0.35 | -0.309 | 0.185 | -0.807 | 0.887 | -0.089 | -0.412 | 0.123 | 0.399 | 0.556 | 0.35 | 0.09 | -2.106 | -1.641 | -0.215 | -1.641 |