Model info
| Transcription factor | Tead2 | ||||||||
| Model | TEAD2_MOUSE.H11DI.0.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 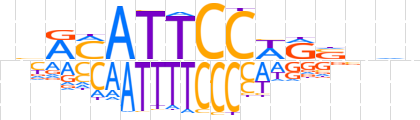 | ||||||||
| LOGO (reverse complement) | 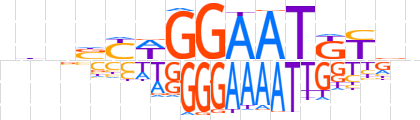 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 15 | ||||||||
| Quality | C | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nbRMATTCCWRSnhb | ||||||||
| Best auROC (human) | 0.551 | ||||||||
| Best auROC (mouse) | 0.946 | ||||||||
| Peak sets in benchmark (human) | 2 | ||||||||
| Peak sets in benchmark (mouse) | 2 | ||||||||
| Aligned words | 309 | ||||||||
| TF family | TEF-1-related factors {3.6.1} | ||||||||
| TF subfamily | TEF-4 (TEAD-2) {3.6.1.0.3} | ||||||||
| MGI | MGI:104904 | ||||||||
| EntrezGene | GeneID:21677 (SSTAR profile) | ||||||||
| UniProt ID | TEAD2_MOUSE | ||||||||
| UniProt AC | P48301 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Tead2 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 9.0 | 25.0 | 18.0 | 15.0 | 29.0 | 55.0 | 3.0 | 32.0 | 5.0 | 15.0 | 14.0 | 15.0 | 9.0 | 32.0 | 21.0 | 11.0 |
| 02 | 34.0 | 1.0 | 17.0 | 0.0 | 116.0 | 1.0 | 8.0 | 2.0 | 36.0 | 1.0 | 19.0 | 0.0 | 14.0 | 0.0 | 59.0 | 0.0 |
| 03 | 30.0 | 147.0 | 4.0 | 19.0 | 0.0 | 1.0 | 1.0 | 1.0 | 28.0 | 73.0 | 1.0 | 1.0 | 0.0 | 2.0 | 0.0 | 0.0 |
| 04 | 55.0 | 1.0 | 2.0 | 0.0 | 219.0 | 2.0 | 1.0 | 1.0 | 5.0 | 1.0 | 0.0 | 0.0 | 21.0 | 0.0 | 0.0 | 0.0 |
| 05 | 2.0 | 8.0 | 2.0 | 288.0 | 0.0 | 1.0 | 0.0 | 3.0 | 1.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 1.0 |
| 06 | 0.0 | 0.0 | 1.0 | 2.0 | 0.0 | 0.0 | 0.0 | 9.0 | 1.0 | 0.0 | 0.0 | 1.0 | 23.0 | 0.0 | 1.0 | 270.0 |
| 07 | 0.0 | 23.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 279.0 | 2.0 | 1.0 |
| 08 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 287.0 | 1.0 | 16.0 | 0.0 | 3.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 |
| 09 | 0.0 | 0.0 | 0.0 | 0.0 | 166.0 | 17.0 | 6.0 | 102.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 16.0 |
| 10 | 28.0 | 3.0 | 130.0 | 6.0 | 11.0 | 2.0 | 1.0 | 3.0 | 3.0 | 1.0 | 1.0 | 1.0 | 9.0 | 7.0 | 89.0 | 13.0 |
| 11 | 4.0 | 12.0 | 30.0 | 5.0 | 2.0 | 6.0 | 3.0 | 2.0 | 29.0 | 40.0 | 130.0 | 22.0 | 0.0 | 4.0 | 19.0 | 0.0 |
| 12 | 8.0 | 7.0 | 18.0 | 2.0 | 26.0 | 16.0 | 3.0 | 17.0 | 36.0 | 60.0 | 56.0 | 30.0 | 4.0 | 7.0 | 11.0 | 7.0 |
| 13 | 29.0 | 21.0 | 17.0 | 7.0 | 50.0 | 9.0 | 7.0 | 24.0 | 35.0 | 24.0 | 13.0 | 16.0 | 4.0 | 31.0 | 12.0 | 9.0 |
| 14 | 17.0 | 10.0 | 29.0 | 62.0 | 19.0 | 36.0 | 5.0 | 25.0 | 12.0 | 12.0 | 12.0 | 13.0 | 4.0 | 10.0 | 18.0 | 24.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.74 | 0.257 | -0.066 | -0.244 | 0.404 | 1.038 | -1.765 | 0.501 | -1.297 | -0.244 | -0.312 | -0.244 | -0.74 | 0.501 | 0.085 | -0.546 |
| 02 | 0.561 | -2.67 | -0.122 | -4.003 | 1.781 | -2.67 | -0.853 | -2.118 | 0.617 | -2.67 | -0.013 | -4.003 | -0.312 | -4.003 | 1.108 | -4.003 |
| 03 | 0.437 | 2.017 | -1.504 | -0.013 | -4.003 | -2.67 | -2.67 | -2.67 | 0.369 | 1.319 | -2.67 | -2.67 | -4.003 | -2.118 | -4.003 | -4.003 |
| 04 | 1.038 | -2.67 | -2.118 | -4.003 | 2.415 | -2.118 | -2.67 | -2.67 | -1.297 | -2.67 | -4.003 | -4.003 | 0.085 | -4.003 | -4.003 | -4.003 |
| 05 | -2.118 | -0.853 | -2.118 | 2.688 | -4.003 | -2.67 | -4.003 | -1.765 | -2.67 | -4.003 | -4.003 | -2.118 | -4.003 | -4.003 | -4.003 | -2.67 |
| 06 | -4.003 | -4.003 | -2.67 | -2.118 | -4.003 | -4.003 | -4.003 | -0.74 | -2.67 | -4.003 | -4.003 | -2.67 | 0.175 | -4.003 | -2.67 | 2.624 |
| 07 | -4.003 | 0.175 | -2.67 | -4.003 | -4.003 | -4.003 | -4.003 | -4.003 | -4.003 | -2.118 | -4.003 | -4.003 | -4.003 | 2.657 | -2.118 | -2.67 |
| 08 | -4.003 | -4.003 | -4.003 | -4.003 | -4.003 | 2.685 | -2.67 | -0.181 | -4.003 | -1.765 | -4.003 | -4.003 | -4.003 | -2.67 | -4.003 | -4.003 |
| 09 | -4.003 | -4.003 | -4.003 | -4.003 | 2.138 | -0.122 | -1.126 | 1.653 | -2.67 | -4.003 | -4.003 | -4.003 | -4.003 | -4.003 | -4.003 | -0.181 |
| 10 | 0.369 | -1.765 | 1.894 | -1.126 | -0.546 | -2.118 | -2.67 | -1.765 | -1.765 | -2.67 | -2.67 | -2.67 | -0.74 | -0.98 | 1.517 | -0.384 |
| 11 | -1.504 | -0.462 | 0.437 | -1.297 | -2.118 | -1.126 | -1.765 | -2.118 | 0.404 | 0.722 | 1.894 | 0.131 | -4.003 | -1.504 | -0.013 | -4.003 |
| 12 | -0.853 | -0.98 | -0.066 | -2.118 | 0.296 | -0.181 | -1.765 | -0.122 | 0.617 | 1.124 | 1.056 | 0.437 | -1.504 | -0.98 | -0.546 | -0.98 |
| 13 | 0.404 | 0.085 | -0.122 | -0.98 | 0.943 | -0.74 | -0.98 | 0.217 | 0.59 | 0.217 | -0.384 | -0.181 | -1.504 | 0.47 | -0.462 | -0.74 |
| 14 | -0.122 | -0.638 | 0.404 | 1.157 | -0.013 | 0.617 | -1.297 | 0.257 | -0.462 | -0.462 | -0.462 | -0.384 | -1.504 | -0.638 | -0.066 | 0.217 |