Model info
| Transcription factor | Tfcp2l1 | ||||||||
| Model | TF2L1_MOUSE.H11DI.0.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 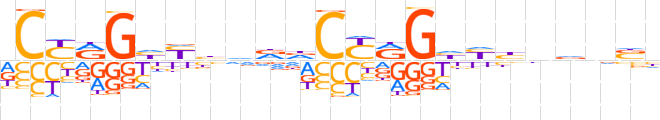 | ||||||||
| LOGO (reverse complement) | 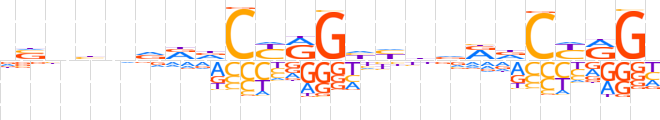 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 23 | ||||||||
| Quality | C | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | dCYRGhYbdddCYRGhYbnvnbh | ||||||||
| Best auROC (human) | |||||||||
| Best auROC (mouse) | 0.995 | ||||||||
| Peak sets in benchmark (human) | |||||||||
| Peak sets in benchmark (mouse) | 3 | ||||||||
| Aligned words | 454 | ||||||||
| TF family | CP2-related factors {6.7.2} | ||||||||
| TF subfamily | CP2-L1 (LBP-9, CRTR-1) {6.7.2.0.2} | ||||||||
| MGI | MGI:2444691 | ||||||||
| EntrezGene | GeneID:81879 (SSTAR profile) | ||||||||
| UniProt ID | TF2L1_MOUSE | ||||||||
| UniProt AC | Q3UNW5 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Tfcp2l1 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.0 | 162.0 | 3.0 | 0.0 | 0.0 | 44.0 | 1.0 | 2.0 | 0.0 | 129.0 | 4.0 | 2.0 | 0.0 | 91.0 | 7.0 | 2.0 |
| 02 | 0.0 | 0.0 | 0.0 | 0.0 | 31.0 | 240.0 | 0.0 | 155.0 | 0.0 | 10.0 | 2.0 | 3.0 | 0.0 | 2.0 | 1.0 | 3.0 |
| 03 | 8.0 | 0.0 | 23.0 | 0.0 | 134.0 | 2.0 | 79.0 | 37.0 | 0.0 | 1.0 | 2.0 | 0.0 | 15.0 | 1.0 | 133.0 | 12.0 |
| 04 | 1.0 | 1.0 | 154.0 | 1.0 | 1.0 | 0.0 | 3.0 | 0.0 | 3.0 | 16.0 | 217.0 | 1.0 | 0.0 | 0.0 | 48.0 | 1.0 |
| 05 | 1.0 | 2.0 | 2.0 | 0.0 | 1.0 | 11.0 | 0.0 | 5.0 | 78.0 | 123.0 | 18.0 | 203.0 | 0.0 | 2.0 | 1.0 | 0.0 |
| 06 | 14.0 | 13.0 | 14.0 | 39.0 | 9.0 | 32.0 | 2.0 | 95.0 | 1.0 | 7.0 | 2.0 | 11.0 | 14.0 | 52.0 | 14.0 | 128.0 |
| 07 | 6.0 | 18.0 | 9.0 | 5.0 | 22.0 | 48.0 | 2.0 | 32.0 | 5.0 | 17.0 | 9.0 | 1.0 | 27.0 | 122.0 | 48.0 | 76.0 |
| 08 | 26.0 | 4.0 | 24.0 | 6.0 | 106.0 | 36.0 | 11.0 | 52.0 | 30.0 | 8.0 | 22.0 | 8.0 | 27.0 | 22.0 | 42.0 | 23.0 |
| 09 | 78.0 | 10.0 | 86.0 | 15.0 | 46.0 | 8.0 | 8.0 | 8.0 | 58.0 | 8.0 | 27.0 | 6.0 | 24.0 | 5.0 | 43.0 | 17.0 |
| 10 | 92.0 | 1.0 | 67.0 | 46.0 | 15.0 | 2.0 | 0.0 | 14.0 | 93.0 | 9.0 | 42.0 | 20.0 | 8.0 | 4.0 | 7.0 | 27.0 |
| 11 | 0.0 | 202.0 | 6.0 | 0.0 | 0.0 | 16.0 | 0.0 | 0.0 | 1.0 | 103.0 | 11.0 | 1.0 | 0.0 | 101.0 | 4.0 | 2.0 |
| 12 | 0.0 | 1.0 | 0.0 | 0.0 | 35.0 | 259.0 | 1.0 | 127.0 | 0.0 | 16.0 | 1.0 | 4.0 | 0.0 | 1.0 | 0.0 | 2.0 |
| 13 | 5.0 | 0.0 | 30.0 | 0.0 | 116.0 | 5.0 | 111.0 | 45.0 | 1.0 | 0.0 | 0.0 | 1.0 | 17.0 | 1.0 | 111.0 | 4.0 |
| 14 | 0.0 | 0.0 | 139.0 | 0.0 | 1.0 | 0.0 | 5.0 | 0.0 | 2.0 | 15.0 | 235.0 | 0.0 | 0.0 | 1.0 | 49.0 | 0.0 |
| 15 | 1.0 | 1.0 | 0.0 | 1.0 | 1.0 | 8.0 | 0.0 | 7.0 | 90.0 | 110.0 | 22.0 | 206.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 16 | 30.0 | 22.0 | 10.0 | 30.0 | 6.0 | 28.0 | 6.0 | 79.0 | 8.0 | 5.0 | 3.0 | 6.0 | 14.0 | 36.0 | 16.0 | 148.0 |
| 17 | 9.0 | 26.0 | 8.0 | 15.0 | 12.0 | 38.0 | 8.0 | 33.0 | 1.0 | 22.0 | 6.0 | 6.0 | 20.0 | 116.0 | 30.0 | 97.0 |
| 18 | 11.0 | 11.0 | 13.0 | 7.0 | 64.0 | 49.0 | 12.0 | 77.0 | 7.0 | 10.0 | 24.0 | 11.0 | 28.0 | 36.0 | 54.0 | 33.0 |
| 19 | 25.0 | 19.0 | 58.0 | 8.0 | 56.0 | 16.0 | 14.0 | 20.0 | 41.0 | 15.0 | 29.0 | 18.0 | 23.0 | 21.0 | 70.0 | 14.0 |
| 20 | 40.0 | 15.0 | 62.0 | 28.0 | 21.0 | 18.0 | 6.0 | 26.0 | 46.0 | 36.0 | 60.0 | 29.0 | 12.0 | 11.0 | 23.0 | 14.0 |
| 21 | 12.0 | 71.0 | 31.0 | 5.0 | 8.0 | 47.0 | 1.0 | 24.0 | 19.0 | 83.0 | 41.0 | 8.0 | 4.0 | 51.0 | 31.0 | 11.0 |
| 22 | 11.0 | 17.0 | 13.0 | 2.0 | 46.0 | 101.0 | 9.0 | 96.0 | 16.0 | 49.0 | 21.0 | 18.0 | 5.0 | 19.0 | 18.0 | 6.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -4.307 | 1.746 | -2.125 | -4.307 | -4.307 | 0.449 | -3.02 | -2.476 | -4.307 | 1.519 | -1.866 | -2.476 | -4.307 | 1.172 | -1.345 | -2.476 |
| 02 | -4.307 | -4.307 | -4.307 | -4.307 | 0.103 | 2.139 | -4.307 | 1.702 | -4.307 | -1.004 | -2.476 | -2.125 | -4.307 | -2.476 | -3.02 | -2.125 |
| 03 | -1.218 | -4.307 | -0.192 | -4.307 | 1.557 | -2.476 | 1.031 | 0.278 | -4.307 | -3.02 | -2.476 | -4.307 | -0.61 | -3.02 | 1.55 | -0.827 |
| 04 | -3.02 | -3.02 | 1.696 | -3.02 | -3.02 | -4.307 | -2.125 | -4.307 | -2.125 | -0.547 | 2.038 | -3.02 | -4.307 | -4.307 | 0.536 | -3.02 |
| 05 | -3.02 | -2.476 | -2.476 | -4.307 | -3.02 | -0.912 | -4.307 | -1.661 | 1.018 | 1.472 | -0.432 | 1.972 | -4.307 | -2.476 | -3.02 | -4.307 |
| 06 | -0.678 | -0.75 | -0.678 | 0.33 | -1.105 | 0.134 | -2.476 | 1.214 | -3.02 | -1.345 | -2.476 | -0.912 | -0.678 | 0.615 | -0.678 | 1.511 |
| 07 | -1.49 | -0.432 | -1.105 | -1.661 | -0.235 | 0.536 | -2.476 | 0.134 | -1.661 | -0.488 | -1.105 | -3.02 | -0.034 | 1.464 | 0.536 | 0.992 |
| 08 | -0.071 | -1.866 | -0.15 | -1.49 | 1.324 | 0.251 | -0.912 | 0.615 | 0.07 | -1.218 | -0.235 | -1.218 | -0.034 | -0.235 | 0.403 | -0.192 |
| 09 | 1.018 | -1.004 | 1.115 | -0.61 | 0.493 | -1.218 | -1.218 | -1.218 | 0.723 | -1.218 | -0.034 | -1.49 | -0.15 | -1.661 | 0.427 | -0.488 |
| 10 | 1.182 | -3.02 | 0.867 | 0.493 | -0.61 | -2.476 | -4.307 | -0.678 | 1.193 | -1.105 | 0.403 | -0.329 | -1.218 | -1.866 | -1.345 | -0.034 |
| 11 | -4.307 | 1.967 | -1.49 | -4.307 | -4.307 | -0.547 | -4.307 | -4.307 | -3.02 | 1.295 | -0.912 | -3.02 | -4.307 | 1.275 | -1.866 | -2.476 |
| 12 | -4.307 | -3.02 | -4.307 | -4.307 | 0.223 | 2.215 | -3.02 | 1.504 | -4.307 | -0.547 | -3.02 | -1.866 | -4.307 | -3.02 | -4.307 | -2.476 |
| 13 | -1.661 | -4.307 | 0.07 | -4.307 | 1.413 | -1.661 | 1.369 | 0.472 | -3.02 | -4.307 | -4.307 | -3.02 | -0.488 | -3.02 | 1.369 | -1.866 |
| 14 | -4.307 | -4.307 | 1.594 | -4.307 | -3.02 | -4.307 | -1.661 | -4.307 | -2.476 | -0.61 | 2.118 | -4.307 | -4.307 | -3.02 | 0.556 | -4.307 |
| 15 | -3.02 | -3.02 | -4.307 | -3.02 | -3.02 | -1.218 | -4.307 | -1.345 | 1.161 | 1.36 | -0.235 | 1.986 | -4.307 | -4.307 | -4.307 | -4.307 |
| 16 | 0.07 | -0.235 | -1.004 | 0.07 | -1.49 | 0.002 | -1.49 | 1.031 | -1.218 | -1.661 | -2.125 | -1.49 | -0.678 | 0.251 | -0.547 | 1.656 |
| 17 | -1.105 | -0.071 | -1.218 | -0.61 | -0.827 | 0.304 | -1.218 | 0.164 | -3.02 | -0.235 | -1.49 | -1.49 | -0.329 | 1.413 | 0.07 | 1.235 |
| 18 | -0.912 | -0.912 | -0.75 | -1.345 | 0.821 | 0.556 | -0.827 | 1.005 | -1.345 | -1.004 | -0.15 | -0.912 | 0.002 | 0.251 | 0.652 | 0.164 |
| 19 | -0.11 | -0.379 | 0.723 | -1.218 | 0.689 | -0.547 | -0.678 | -0.329 | 0.379 | -0.61 | 0.037 | -0.432 | -0.192 | -0.281 | 0.91 | -0.678 |
| 20 | 0.355 | -0.61 | 0.79 | 0.002 | -0.281 | -0.432 | -1.49 | -0.071 | 0.493 | 0.251 | 0.757 | 0.037 | -0.827 | -0.912 | -0.192 | -0.678 |
| 21 | -0.827 | 0.925 | 0.103 | -1.661 | -1.218 | 0.515 | -3.02 | -0.15 | -0.379 | 1.08 | 0.379 | -1.218 | -1.866 | 0.596 | 0.103 | -0.912 |
| 22 | -0.912 | -0.488 | -0.75 | -2.476 | 0.493 | 1.275 | -1.105 | 1.225 | -0.547 | 0.556 | -0.281 | -0.432 | -1.661 | -0.379 | -0.432 | -1.49 |