Model info
| Transcription factor | RELA (GeneCards) | ||||||||
| Model | TF65_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 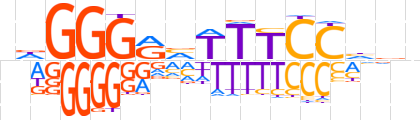 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 15 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | ndGGGRvTTTCChvn | ||||||||
| Best auROC (human) | 0.988 | ||||||||
| Best auROC (mouse) | 0.91 | ||||||||
| Peak sets in benchmark (human) | 188 | ||||||||
| Peak sets in benchmark (mouse) | 99 | ||||||||
| Aligned words | 504 | ||||||||
| TF family | NF-kappaB-related factors {6.1.1} | ||||||||
| TF subfamily | NF-kappaB p65 subunit-like factors {6.1.1.2} | ||||||||
| HGNC | HGNC:9955 | ||||||||
| EntrezGene | GeneID:5970 (SSTAR profile) | ||||||||
| UniProt ID | TF65_HUMAN | ||||||||
| UniProt AC | Q04206 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | RELA expression | ||||||||
| ReMap ChIP-seq dataset list | RELA datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 68.0 | 3.0 | 34.0 | 24.0 | 71.0 | 5.0 | 11.0 | 40.0 | 44.0 | 5.0 | 15.0 | 24.0 | 57.0 | 4.0 | 59.0 | 35.0 |
| 02 | 0.0 | 1.0 | 236.0 | 3.0 | 0.0 | 0.0 | 16.0 | 1.0 | 0.0 | 1.0 | 118.0 | 0.0 | 0.0 | 0.0 | 123.0 | 0.0 |
| 03 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 3.0 | 1.0 | 483.0 | 6.0 | 1.0 | 1.0 | 2.0 | 0.0 |
| 04 | 2.0 | 0.0 | 1.0 | 1.0 | 1.0 | 0.0 | 0.0 | 1.0 | 1.0 | 1.0 | 450.0 | 35.0 | 1.0 | 0.0 | 3.0 | 2.0 |
| 05 | 3.0 | 0.0 | 2.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 185.0 | 2.0 | 251.0 | 16.0 | 10.0 | 0.0 | 26.0 | 3.0 |
| 06 | 73.0 | 96.0 | 24.0 | 6.0 | 1.0 | 0.0 | 0.0 | 1.0 | 178.0 | 50.0 | 31.0 | 20.0 | 10.0 | 4.0 | 1.0 | 4.0 |
| 07 | 102.0 | 7.0 | 5.0 | 148.0 | 5.0 | 2.0 | 0.0 | 143.0 | 4.0 | 1.0 | 0.0 | 51.0 | 2.0 | 0.0 | 0.0 | 29.0 |
| 08 | 22.0 | 10.0 | 8.0 | 73.0 | 1.0 | 0.0 | 1.0 | 8.0 | 1.0 | 0.0 | 0.0 | 4.0 | 0.0 | 7.0 | 7.0 | 357.0 |
| 09 | 3.0 | 12.0 | 0.0 | 9.0 | 0.0 | 1.0 | 0.0 | 16.0 | 2.0 | 6.0 | 1.0 | 7.0 | 8.0 | 64.0 | 5.0 | 365.0 |
| 10 | 0.0 | 13.0 | 0.0 | 0.0 | 3.0 | 80.0 | 0.0 | 0.0 | 0.0 | 5.0 | 1.0 | 0.0 | 11.0 | 354.0 | 1.0 | 31.0 |
| 11 | 0.0 | 14.0 | 0.0 | 0.0 | 21.0 | 401.0 | 1.0 | 29.0 | 0.0 | 1.0 | 1.0 | 0.0 | 1.0 | 15.0 | 2.0 | 13.0 |
| 12 | 5.0 | 9.0 | 4.0 | 4.0 | 212.0 | 131.0 | 25.0 | 63.0 | 3.0 | 0.0 | 0.0 | 1.0 | 4.0 | 10.0 | 18.0 | 10.0 |
| 13 | 55.0 | 46.0 | 100.0 | 23.0 | 93.0 | 31.0 | 10.0 | 16.0 | 7.0 | 11.0 | 21.0 | 8.0 | 10.0 | 17.0 | 37.0 | 14.0 |
| 14 | 46.0 | 26.0 | 77.0 | 16.0 | 39.0 | 14.0 | 14.0 | 38.0 | 49.0 | 41.0 | 49.0 | 29.0 | 13.0 | 9.0 | 26.0 | 13.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.773 | -2.232 | 0.085 | -0.258 | 0.816 | -1.768 | -1.02 | 0.246 | 0.341 | -1.768 | -0.719 | -0.258 | 0.597 | -1.973 | 0.632 | 0.114 |
| 02 | -4.398 | -3.124 | 2.013 | -2.232 | -4.398 | -4.398 | -0.656 | -3.124 | -4.398 | -3.124 | 1.322 | -4.398 | -4.398 | -4.398 | 1.363 | -4.398 |
| 03 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -2.582 | -4.398 | -2.232 | -3.124 | 2.728 | -1.598 | -3.124 | -3.124 | -2.582 | -4.398 |
| 04 | -2.582 | -4.398 | -3.124 | -3.124 | -3.124 | -4.398 | -4.398 | -3.124 | -3.124 | -3.124 | 2.658 | 0.114 | -3.124 | -4.398 | -2.232 | -2.582 |
| 05 | -2.232 | -4.398 | -2.582 | -4.398 | -3.124 | -4.398 | -4.398 | -4.398 | 1.77 | -2.582 | 2.075 | -0.656 | -1.112 | -4.398 | -0.179 | -2.232 |
| 06 | 0.843 | 1.116 | -0.258 | -1.598 | -3.124 | -4.398 | -4.398 | -3.124 | 1.732 | 0.467 | -0.006 | -0.437 | -1.112 | -1.973 | -3.124 | -1.973 |
| 07 | 1.176 | -1.452 | -1.768 | 1.547 | -1.768 | -2.582 | -4.398 | 1.513 | -1.973 | -3.124 | -4.398 | 0.487 | -2.582 | -4.398 | -4.398 | -0.072 |
| 08 | -0.344 | -1.112 | -1.326 | 0.843 | -3.124 | -4.398 | -3.124 | -1.326 | -3.124 | -4.398 | -4.398 | -1.973 | -4.398 | -1.452 | -1.452 | 2.426 |
| 09 | -2.232 | -0.936 | -4.398 | -1.213 | -4.398 | -3.124 | -4.398 | -0.656 | -2.582 | -1.598 | -3.124 | -1.452 | -1.326 | 0.713 | -1.768 | 2.449 |
| 10 | -4.398 | -0.858 | -4.398 | -4.398 | -2.232 | 0.934 | -4.398 | -4.398 | -4.398 | -1.768 | -3.124 | -4.398 | -1.02 | 2.418 | -3.124 | -0.006 |
| 11 | -4.398 | -0.786 | -4.398 | -4.398 | -0.39 | 2.543 | -3.124 | -0.072 | -4.398 | -3.124 | -3.124 | -4.398 | -3.124 | -0.719 | -2.582 | -0.858 |
| 12 | -1.768 | -1.213 | -1.973 | -1.973 | 1.906 | 1.426 | -0.218 | 0.697 | -2.232 | -4.398 | -4.398 | -3.124 | -1.973 | -1.112 | -0.541 | -1.112 |
| 13 | 0.562 | 0.385 | 1.157 | -0.3 | 1.084 | -0.006 | -1.112 | -0.656 | -1.452 | -1.02 | -0.39 | -1.326 | -1.112 | -0.597 | 0.169 | -0.786 |
| 14 | 0.385 | -0.179 | 0.896 | -0.656 | 0.221 | -0.786 | -0.786 | 0.195 | 0.447 | 0.271 | 0.447 | -0.072 | -0.858 | -1.213 | -0.179 | -0.858 |