Model info
| Transcription factor | Tcf7l1 | ||||||||
| Model | TF7L1_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 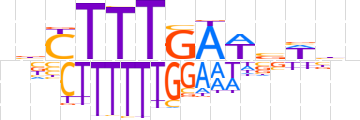 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 13 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nbCTTTGAWvWbn | ||||||||
| Best auROC (human) | 0.864 | ||||||||
| Best auROC (mouse) | 0.861 | ||||||||
| Peak sets in benchmark (human) | 10 | ||||||||
| Peak sets in benchmark (mouse) | 10 | ||||||||
| Aligned words | 222 | ||||||||
| TF family | TCF-7-related factors {4.1.3} | ||||||||
| TF subfamily | TCF-7L1 (TCF-3) [1] {4.1.3.0.2} | ||||||||
| MGI | MGI:1202876 | ||||||||
| EntrezGene | GeneID:21415 (SSTAR profile) | ||||||||
| UniProt ID | TF7L1_MOUSE | ||||||||
| UniProt AC | Q9Z1J1 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Tcf7l1 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 3.0 | 15.0 | 9.0 | 12.0 | 7.0 | 22.0 | 7.0 | 27.0 | 4.0 | 20.0 | 18.0 | 10.0 | 5.0 | 21.0 | 24.0 | 18.0 |
| 02 | 0.0 | 18.0 | 1.0 | 0.0 | 0.0 | 59.0 | 1.0 | 18.0 | 0.0 | 50.0 | 1.0 | 7.0 | 1.0 | 48.0 | 8.0 | 10.0 |
| 03 | 0.0 | 0.0 | 0.0 | 1.0 | 1.0 | 2.0 | 0.0 | 172.0 | 0.0 | 0.0 | 0.0 | 11.0 | 0.0 | 0.0 | 0.0 | 35.0 |
| 04 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 5.0 | 1.0 | 213.0 |
| 05 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 6.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 214.0 |
| 06 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 6.0 | 34.0 | 180.0 | 1.0 |
| 07 | 5.0 | 0.0 | 0.0 | 1.0 | 29.0 | 0.0 | 0.0 | 5.0 | 154.0 | 0.0 | 7.0 | 20.0 | 1.0 | 0.0 | 0.0 | 0.0 |
| 08 | 80.0 | 0.0 | 2.0 | 107.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.0 | 2.0 | 0.0 | 2.0 | 5.0 | 0.0 | 2.0 | 19.0 |
| 09 | 12.0 | 19.0 | 53.0 | 4.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 3.0 | 0.0 | 14.0 | 53.0 | 43.0 | 18.0 |
| 10 | 13.0 | 8.0 | 1.0 | 5.0 | 10.0 | 7.0 | 4.0 | 52.0 | 12.0 | 7.0 | 8.0 | 73.0 | 1.0 | 3.0 | 3.0 | 15.0 |
| 11 | 8.0 | 10.0 | 6.0 | 12.0 | 3.0 | 7.0 | 3.0 | 12.0 | 2.0 | 7.0 | 3.0 | 4.0 | 11.0 | 56.0 | 37.0 | 41.0 |
| 12 | 8.0 | 3.0 | 8.0 | 5.0 | 18.0 | 25.0 | 9.0 | 28.0 | 6.0 | 14.0 | 23.0 | 6.0 | 11.0 | 19.0 | 20.0 | 19.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -1.449 | 0.076 | -0.42 | -0.141 | -0.661 | 0.452 | -0.661 | 0.654 | -1.187 | 0.358 | 0.255 | -0.318 | -0.979 | 0.406 | 0.538 | 0.255 |
| 02 | -3.74 | 0.255 | -2.363 | -3.74 | -3.74 | 1.429 | -2.363 | 0.255 | -3.74 | 1.265 | -2.363 | -0.661 | -2.363 | 1.224 | -0.533 | -0.318 |
| 03 | -3.74 | -3.74 | -3.74 | -2.363 | -2.363 | -1.805 | -3.74 | 2.495 | -3.74 | -3.74 | -3.74 | -0.226 | -3.74 | -3.74 | -3.74 | 0.911 |
| 04 | -3.74 | -3.74 | -3.74 | -2.363 | -3.74 | -2.363 | -3.74 | -2.363 | -3.74 | -3.74 | -3.74 | -3.74 | -3.74 | -0.979 | -2.363 | 2.709 |
| 05 | -3.74 | -3.74 | -3.74 | -3.74 | -3.74 | -3.74 | -3.74 | -0.808 | -3.74 | -3.74 | -3.74 | -2.363 | -3.74 | -2.363 | -3.74 | 2.713 |
| 06 | -3.74 | -3.74 | -3.74 | -3.74 | -3.74 | -3.74 | -2.363 | -3.74 | -3.74 | -3.74 | -3.74 | -3.74 | -0.808 | 0.882 | 2.541 | -2.363 |
| 07 | -0.979 | -3.74 | -3.74 | -2.363 | 0.725 | -3.74 | -3.74 | -0.979 | 2.385 | -3.74 | -0.661 | 0.358 | -2.363 | -3.74 | -3.74 | -3.74 |
| 08 | 1.732 | -3.74 | -1.805 | 2.022 | -3.74 | -3.74 | -3.74 | -3.74 | -1.449 | -1.805 | -3.74 | -1.805 | -0.979 | -3.74 | -1.805 | 0.308 |
| 09 | -0.141 | 0.308 | 1.323 | -1.187 | -2.363 | -3.74 | -2.363 | -3.74 | -3.74 | -2.363 | -1.449 | -3.74 | 0.009 | 1.323 | 1.115 | 0.255 |
| 10 | -0.064 | -0.533 | -2.363 | -0.979 | -0.318 | -0.661 | -1.187 | 1.304 | -0.141 | -0.661 | -0.533 | 1.641 | -2.363 | -1.449 | -1.449 | 0.076 |
| 11 | -0.533 | -0.318 | -0.808 | -0.141 | -1.449 | -0.661 | -1.449 | -0.141 | -1.805 | -0.661 | -1.449 | -1.187 | -0.226 | 1.377 | 0.966 | 1.068 |
| 12 | -0.533 | -1.449 | -0.533 | -0.979 | 0.255 | 0.578 | -0.42 | 0.69 | -0.808 | 0.009 | 0.496 | -0.808 | -0.226 | 0.308 | 0.358 | 0.308 |