Model info
| Transcription factor | TFE3 (GeneCards) | ||||||||
| Model | TFE3_HUMAN.H11DI.0.B | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 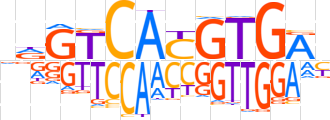 | ||||||||
| LOGO (reverse complement) | 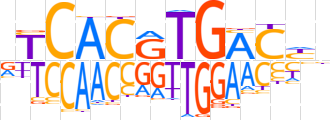 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 12 | ||||||||
| Quality | B | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | ndGTCAYGTGAb | ||||||||
| Best auROC (human) | 0.503 | ||||||||
| Best auROC (mouse) | 1.0 | ||||||||
| Peak sets in benchmark (human) | 2 | ||||||||
| Peak sets in benchmark (mouse) | 30 | ||||||||
| Aligned words | 437 | ||||||||
| TF family | bHLH-ZIP factors {1.2.6} | ||||||||
| TF subfamily | TFE3-like factors {1.2.6.1} | ||||||||
| HGNC | HGNC:11752 | ||||||||
| EntrezGene | GeneID:7030 (SSTAR profile) | ||||||||
| UniProt ID | TFE3_HUMAN | ||||||||
| UniProt AC | P19532 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | TFE3 expression | ||||||||
| ReMap ChIP-seq dataset list | TFE3 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 44.0 | 0.0 | 60.0 | 10.0 | 25.0 | 14.0 | 26.0 | 20.0 | 38.0 | 2.0 | 66.0 | 22.0 | 22.0 | 0.0 | 49.0 | 7.0 |
| 02 | 9.0 | 2.0 | 117.0 | 1.0 | 6.0 | 0.0 | 10.0 | 0.0 | 42.0 | 5.0 | 152.0 | 2.0 | 4.0 | 0.0 | 55.0 | 0.0 |
| 03 | 4.0 | 3.0 | 10.0 | 44.0 | 3.0 | 0.0 | 0.0 | 4.0 | 10.0 | 13.0 | 30.0 | 281.0 | 1.0 | 0.0 | 1.0 | 1.0 |
| 04 | 0.0 | 18.0 | 0.0 | 0.0 | 0.0 | 16.0 | 0.0 | 0.0 | 0.0 | 41.0 | 0.0 | 0.0 | 0.0 | 329.0 | 0.0 | 1.0 |
| 05 | 0.0 | 0.0 | 0.0 | 0.0 | 382.0 | 2.0 | 1.0 | 19.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 |
| 06 | 0.0 | 268.0 | 12.0 | 102.0 | 0.0 | 1.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 12.0 | 2.0 | 5.0 |
| 07 | 0.0 | 0.0 | 0.0 | 0.0 | 18.0 | 6.0 | 256.0 | 1.0 | 0.0 | 5.0 | 8.0 | 1.0 | 4.0 | 1.0 | 105.0 | 0.0 |
| 08 | 1.0 | 2.0 | 1.0 | 18.0 | 0.0 | 1.0 | 0.0 | 11.0 | 11.0 | 5.0 | 11.0 | 342.0 | 0.0 | 0.0 | 0.0 | 2.0 |
| 09 | 0.0 | 1.0 | 11.0 | 0.0 | 0.0 | 0.0 | 7.0 | 1.0 | 0.0 | 0.0 | 12.0 | 0.0 | 1.0 | 0.0 | 371.0 | 1.0 |
| 10 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 322.0 | 31.0 | 39.0 | 9.0 | 2.0 | 0.0 | 0.0 | 0.0 |
| 11 | 4.0 | 155.0 | 44.0 | 121.0 | 8.0 | 4.0 | 6.0 | 15.0 | 7.0 | 8.0 | 14.0 | 10.0 | 0.0 | 3.0 | 5.0 | 1.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.547 | -4.226 | 0.855 | -0.907 | -0.012 | -0.581 | 0.026 | -0.232 | 0.401 | -2.381 | 0.949 | -0.138 | -0.138 | -4.226 | 0.653 | -1.248 |
| 02 | -1.008 | -2.381 | 1.519 | -2.927 | -1.394 | -4.226 | -0.907 | -4.226 | 0.501 | -1.564 | 1.78 | -2.381 | -1.77 | -4.226 | 0.768 | -4.226 |
| 03 | -1.77 | -2.03 | -0.907 | 0.547 | -2.03 | -4.226 | -4.226 | -1.77 | -0.907 | -0.653 | 0.168 | 2.394 | -2.927 | -4.226 | -2.927 | -2.927 |
| 04 | -4.226 | -0.335 | -4.226 | -4.226 | -4.226 | -0.45 | -4.226 | -4.226 | -4.226 | 0.477 | -4.226 | -4.226 | -4.226 | 2.551 | -4.226 | -2.927 |
| 05 | -4.226 | -4.226 | -4.226 | -4.226 | 2.7 | -2.381 | -2.927 | -0.282 | -4.226 | -4.226 | -4.226 | -4.226 | -4.226 | -2.927 | -4.226 | -4.226 |
| 06 | -4.226 | 2.346 | -0.73 | 1.383 | -4.226 | -2.927 | -4.226 | -2.381 | -4.226 | -4.226 | -4.226 | -2.927 | -4.226 | -0.73 | -2.381 | -1.564 |
| 07 | -4.226 | -4.226 | -4.226 | -4.226 | -0.335 | -1.394 | 2.301 | -2.927 | -4.226 | -1.564 | -1.121 | -2.927 | -1.77 | -2.927 | 1.412 | -4.226 |
| 08 | -2.927 | -2.381 | -2.927 | -0.335 | -4.226 | -2.927 | -4.226 | -0.815 | -0.815 | -1.564 | -0.815 | 2.59 | -4.226 | -4.226 | -4.226 | -2.381 |
| 09 | -4.226 | -2.927 | -0.815 | -4.226 | -4.226 | -4.226 | -1.248 | -2.927 | -4.226 | -4.226 | -0.73 | -4.226 | -2.927 | -4.226 | 2.671 | -2.927 |
| 10 | -4.226 | -2.927 | -4.226 | -4.226 | -4.226 | -2.927 | -4.226 | -4.226 | 2.53 | 0.2 | 0.427 | -1.008 | -2.381 | -4.226 | -4.226 | -4.226 |
| 11 | -1.77 | 1.8 | 0.547 | 1.553 | -1.121 | -1.77 | -1.394 | -0.513 | -1.248 | -1.121 | -0.581 | -0.907 | -4.226 | -2.03 | -1.564 | -2.927 |