Model info
| Transcription factor | THRB (GeneCards) | ||||||||
| Model | THB_HUMAN.H11MO.0.C | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO | 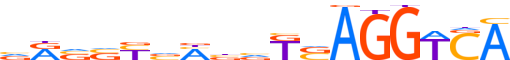 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | Integrative | ||||||||
| Model release | HOCOMOCOv9 | ||||||||
| Model length | 17 | ||||||||
| Quality | C | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | vRvSYvMbvKSAGGTCA | ||||||||
| Best auROC (human) | |||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | |||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 38 | ||||||||
| TF family | Thyroid hormone receptor-related factors (NR1) {2.1.2} | ||||||||
| TF subfamily | Thyroid hormone receptors (NR1A) {2.1.2.2} | ||||||||
| HGNC | HGNC:11799 | ||||||||
| EntrezGene | GeneID:7068 (SSTAR profile) | ||||||||
| UniProt ID | THB_HUMAN | ||||||||
| UniProt AC | P10828 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | THRB expression | ||||||||
| ReMap ChIP-seq dataset list | THRB datasets | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 8.014 | 6.826 | 17.749 | 2.909 |
| 02 | 20.183 | 0.0 | 11.041 | 4.274 |
| 03 | 5.461 | 10.329 | 18.046 | 1.662 |
| 04 | 3.918 | 10.804 | 18.639 | 2.137 |
| 05 | 0.0 | 10.329 | 4.986 | 20.183 |
| 06 | 5.105 | 17.215 | 9.854 | 3.324 |
| 07 | 23.507 | 4.274 | 3.799 | 3.918 |
| 08 | 2.731 | 12.11 | 14.603 | 6.055 |
| 09 | 11.279 | 8.904 | 13.653 | 1.662 |
| 10 | 0.0 | 2.137 | 8.311 | 25.05 |
| 11 | 3.324 | 16.265 | 14.84 | 1.068 |
| 12 | 34.429 | 0.0 | 1.068 | 0.0 |
| 13 | 0.0 | 0.0 | 32.886 | 2.612 |
| 14 | 0.0 | 0.0 | 34.429 | 1.068 |
| 15 | 9.854 | 0.0 | 0.831 | 24.813 |
| 16 | 1.9 | 29.087 | 3.443 | 1.068 |
| 17 | 30.393 | 5.105 | 0.0 | 0.0 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.092 | -0.235 | 0.646 | -0.944 |
| 02 | 0.769 | -2.393 | 0.2 | -0.637 |
| 03 | -0.43 | 0.139 | 0.662 | -1.341 |
| 04 | -0.708 | 0.18 | 0.693 | -1.171 |
| 05 | -2.393 | 0.139 | -0.508 | 0.769 |
| 06 | -0.488 | 0.617 | 0.096 | -0.84 |
| 07 | 0.916 | -0.637 | -0.733 | -0.708 |
| 08 | -0.992 | 0.286 | 0.462 | -0.341 |
| 09 | 0.22 | 0.003 | 0.398 | -1.341 |
| 10 | -2.393 | -1.171 | -0.059 | 0.977 |
| 11 | -0.84 | 0.563 | 0.477 | -1.606 |
| 12 | 1.286 | -2.393 | -1.606 | -2.393 |
| 13 | -2.393 | -2.393 | 1.241 | -1.025 |
| 14 | -2.393 | -2.393 | 1.286 | -1.606 |
| 15 | 0.096 | -2.393 | -1.735 | 0.968 |
| 16 | -1.252 | 1.122 | -0.812 | -1.606 |
| 17 | 1.164 | -0.488 | -2.393 | -2.393 |