Model info
| Transcription factor | Twist1 | ||||||||
| Model | TWST1_MOUSE.H11DI.0.B | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 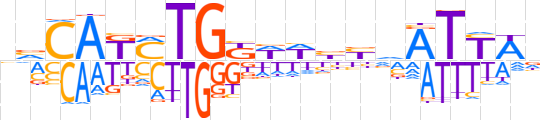 | ||||||||
| LOGO (reverse complement) | 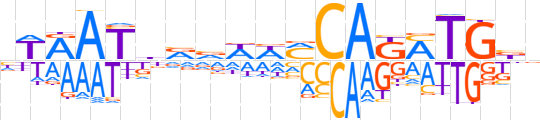 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 19 | ||||||||
| Quality | B | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nvCAKMTGKWWbbvATYWn | ||||||||
| Best auROC (human) | 0.971 | ||||||||
| Best auROC (mouse) | 0.657 | ||||||||
| Peak sets in benchmark (human) | 9 | ||||||||
| Peak sets in benchmark (mouse) | 8 | ||||||||
| Aligned words | 415 | ||||||||
| TF family | Tal-related factors {1.2.3} | ||||||||
| TF subfamily | Twist-like factors {1.2.3.2} | ||||||||
| MGI | MGI:98872 | ||||||||
| EntrezGene | GeneID:22160 (SSTAR profile) | ||||||||
| UniProt ID | TWST1_MOUSE | ||||||||
| UniProt AC | P26687 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Twist1 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 54.0 | 38.0 | 26.0 | 3.0 | 84.0 | 27.0 | 9.0 | 5.0 | 40.0 | 27.0 | 20.0 | 9.0 | 21.0 | 27.0 | 17.0 | 6.0 |
| 02 | 11.0 | 179.0 | 9.0 | 0.0 | 4.0 | 113.0 | 1.0 | 1.0 | 5.0 | 60.0 | 3.0 | 4.0 | 2.0 | 18.0 | 0.0 | 3.0 |
| 03 | 19.0 | 1.0 | 2.0 | 0.0 | 359.0 | 2.0 | 5.0 | 4.0 | 5.0 | 3.0 | 3.0 | 2.0 | 0.0 | 0.0 | 3.0 | 5.0 |
| 04 | 3.0 | 21.0 | 92.0 | 267.0 | 1.0 | 0.0 | 0.0 | 5.0 | 0.0 | 1.0 | 2.0 | 10.0 | 0.0 | 3.0 | 1.0 | 7.0 |
| 05 | 0.0 | 4.0 | 0.0 | 0.0 | 11.0 | 11.0 | 2.0 | 1.0 | 28.0 | 62.0 | 5.0 | 0.0 | 47.0 | 211.0 | 30.0 | 1.0 |
| 06 | 0.0 | 0.0 | 0.0 | 86.0 | 3.0 | 0.0 | 0.0 | 285.0 | 0.0 | 2.0 | 0.0 | 35.0 | 0.0 | 0.0 | 0.0 | 2.0 |
| 07 | 0.0 | 1.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 405.0 | 3.0 |
| 08 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 6.0 | 42.0 | 246.0 | 113.0 | 0.0 | 0.0 | 4.0 | 1.0 |
| 09 | 4.0 | 0.0 | 0.0 | 2.0 | 10.0 | 0.0 | 1.0 | 32.0 | 91.0 | 23.0 | 23.0 | 113.0 | 6.0 | 4.0 | 6.0 | 98.0 |
| 10 | 67.0 | 6.0 | 9.0 | 29.0 | 8.0 | 1.0 | 0.0 | 18.0 | 16.0 | 3.0 | 2.0 | 9.0 | 35.0 | 19.0 | 5.0 | 186.0 |
| 11 | 30.0 | 27.0 | 29.0 | 40.0 | 1.0 | 17.0 | 1.0 | 10.0 | 2.0 | 5.0 | 1.0 | 8.0 | 4.0 | 85.0 | 17.0 | 136.0 |
| 12 | 5.0 | 6.0 | 4.0 | 22.0 | 9.0 | 44.0 | 5.0 | 76.0 | 1.0 | 17.0 | 4.0 | 26.0 | 9.0 | 60.0 | 31.0 | 94.0 |
| 13 | 10.0 | 5.0 | 6.0 | 3.0 | 72.0 | 33.0 | 6.0 | 16.0 | 15.0 | 11.0 | 9.0 | 9.0 | 75.0 | 53.0 | 65.0 | 25.0 |
| 14 | 149.0 | 14.0 | 5.0 | 4.0 | 96.0 | 1.0 | 1.0 | 4.0 | 65.0 | 5.0 | 7.0 | 9.0 | 42.0 | 0.0 | 10.0 | 1.0 |
| 15 | 3.0 | 2.0 | 9.0 | 338.0 | 0.0 | 0.0 | 0.0 | 20.0 | 1.0 | 0.0 | 1.0 | 21.0 | 0.0 | 1.0 | 2.0 | 15.0 |
| 16 | 0.0 | 1.0 | 1.0 | 2.0 | 1.0 | 2.0 | 0.0 | 0.0 | 6.0 | 2.0 | 2.0 | 2.0 | 22.0 | 55.0 | 7.0 | 310.0 |
| 17 | 16.0 | 0.0 | 2.0 | 11.0 | 42.0 | 3.0 | 1.0 | 14.0 | 5.0 | 2.0 | 1.0 | 2.0 | 229.0 | 9.0 | 10.0 | 66.0 |
| 18 | 91.0 | 45.0 | 92.0 | 64.0 | 4.0 | 4.0 | 1.0 | 5.0 | 8.0 | 3.0 | 0.0 | 3.0 | 23.0 | 18.0 | 30.0 | 22.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.731 | 0.382 | 0.007 | -2.049 | 1.17 | 0.044 | -1.027 | -1.583 | 0.433 | 0.044 | -0.251 | -1.027 | -0.203 | 0.044 | -0.41 | -1.413 |
| 02 | -0.834 | 1.924 | -1.027 | -4.242 | -1.789 | 1.465 | -2.946 | -2.946 | -1.583 | 0.835 | -2.049 | -1.789 | -2.4 | -0.354 | -4.242 | -2.049 |
| 03 | -0.301 | -2.946 | -2.4 | -4.242 | 2.619 | -2.4 | -1.583 | -1.789 | -1.583 | -2.049 | -2.049 | -2.4 | -4.242 | -4.242 | -2.049 | -1.583 |
| 04 | -2.049 | -0.203 | 1.261 | 2.323 | -2.946 | -4.242 | -4.242 | -1.583 | -4.242 | -2.946 | -2.4 | -0.926 | -4.242 | -2.049 | -2.946 | -1.267 |
| 05 | -4.242 | -1.789 | -4.242 | -4.242 | -0.834 | -0.834 | -2.4 | -2.946 | 0.08 | 0.868 | -1.583 | -4.242 | 0.593 | 2.088 | 0.148 | -2.946 |
| 06 | -4.242 | -4.242 | -4.242 | 1.193 | -2.049 | -4.242 | -4.242 | 2.388 | -4.242 | -2.4 | -4.242 | 0.301 | -4.242 | -4.242 | -4.242 | -2.4 |
| 07 | -4.242 | -2.946 | -2.4 | -4.242 | -4.242 | -4.242 | -4.242 | -2.4 | -4.242 | -4.242 | -4.242 | -4.242 | -4.242 | -4.242 | 2.739 | -2.049 |
| 08 | -4.242 | -4.242 | -4.242 | -4.242 | -4.242 | -2.946 | -4.242 | -4.242 | -1.413 | 0.481 | 2.242 | 1.465 | -4.242 | -4.242 | -1.789 | -2.946 |
| 09 | -1.789 | -4.242 | -4.242 | -2.4 | -0.926 | -4.242 | -2.946 | 0.212 | 1.25 | -0.114 | -0.114 | 1.465 | -1.413 | -1.789 | -1.413 | 1.323 |
| 10 | 0.945 | -1.413 | -1.027 | 0.115 | -1.14 | -2.946 | -4.242 | -0.354 | -0.469 | -2.049 | -2.4 | -1.027 | 0.301 | -0.301 | -1.583 | 1.962 |
| 11 | 0.148 | 0.044 | 0.115 | 0.433 | -2.946 | -0.41 | -2.946 | -0.926 | -2.4 | -1.583 | -2.946 | -1.14 | -1.789 | 1.182 | -0.41 | 1.65 |
| 12 | -1.583 | -1.413 | -1.789 | -0.157 | -1.027 | 0.527 | -1.583 | 1.07 | -2.946 | -0.41 | -1.789 | 0.007 | -1.027 | 0.835 | 0.181 | 1.282 |
| 13 | -0.926 | -1.583 | -1.413 | -2.049 | 1.017 | 0.243 | -1.413 | -0.469 | -0.533 | -0.834 | -1.027 | -1.027 | 1.057 | 0.712 | 0.915 | -0.032 |
| 14 | 1.741 | -0.6 | -1.583 | -1.789 | 1.303 | -2.946 | -2.946 | -1.789 | 0.915 | -1.583 | -1.267 | -1.027 | 0.481 | -4.242 | -0.926 | -2.946 |
| 15 | -2.049 | -2.4 | -1.027 | 2.559 | -4.242 | -4.242 | -4.242 | -0.251 | -2.946 | -4.242 | -2.946 | -0.203 | -4.242 | -2.946 | -2.4 | -0.533 |
| 16 | -4.242 | -2.946 | -2.946 | -2.4 | -2.946 | -2.4 | -4.242 | -4.242 | -1.413 | -2.4 | -2.4 | -2.4 | -0.157 | 0.749 | -1.267 | 2.472 |
| 17 | -0.469 | -4.242 | -2.4 | -0.834 | 0.481 | -2.049 | -2.946 | -0.6 | -1.583 | -2.4 | -2.946 | -2.4 | 2.17 | -1.027 | -0.926 | 0.93 |
| 18 | 1.25 | 0.55 | 1.261 | 0.899 | -1.789 | -1.789 | -2.946 | -1.583 | -1.14 | -2.049 | -4.242 | -2.049 | -0.114 | -0.354 | 0.148 | -0.157 |