Model info
| Transcription factor | YY1 (GeneCards) | ||||||||
| Model | TYY1_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 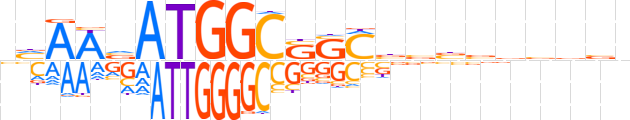 | ||||||||
| LOGO (reverse complement) | 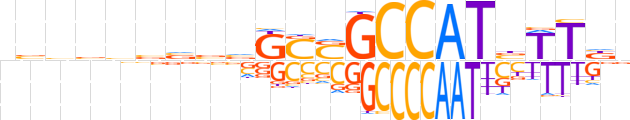 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv10 | ||||||||
| Model length | 22 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nvAAvATGGCSSCbvbvvbvbn | ||||||||
| Best auROC (human) | 0.993 | ||||||||
| Best auROC (mouse) | 0.997 | ||||||||
| Peak sets in benchmark (human) | 69 | ||||||||
| Peak sets in benchmark (mouse) | 72 | ||||||||
| Aligned words | 505 | ||||||||
| TF family | More than 3 adjacent zinc finger factors {2.3.3} | ||||||||
| TF subfamily | YY1-like factors {2.3.3.9} | ||||||||
| HGNC | HGNC:12856 | ||||||||
| EntrezGene | GeneID:7528 (SSTAR profile) | ||||||||
| UniProt ID | TYY1_HUMAN | ||||||||
| UniProt AC | P25490 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | YY1 expression | ||||||||
| ReMap ChIP-seq dataset list | YY1 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 11.789 | 47.192 | 18.386 | 5.982 | 44.45 | 117.046 | 6.281 | 25.162 | 22.289 | 57.86 | 31.922 | 9.885 | 18.921 | 63.613 | 8.455 | 9.929 |
| 02 | 82.829 | 0.844 | 13.776 | 0.0 | 264.142 | 3.034 | 16.38 | 2.155 | 55.435 | 1.825 | 6.121 | 1.663 | 37.462 | 2.962 | 10.534 | 0.0 |
| 03 | 396.212 | 8.413 | 13.284 | 21.96 | 0.0 | 1.884 | 1.858 | 4.924 | 10.878 | 14.66 | 8.67 | 12.603 | 0.847 | 2.155 | 0.0 | 0.816 |
| 04 | 90.365 | 73.911 | 223.861 | 19.8 | 12.987 | 4.221 | 7.987 | 1.917 | 2.77 | 11.781 | 3.546 | 5.715 | 6.659 | 28.66 | 2.903 | 2.081 |
| 05 | 112.781 | 0.0 | 0.0 | 0.0 | 117.3 | 0.0 | 1.273 | 0.0 | 236.655 | 0.833 | 0.0 | 0.808 | 29.512 | 0.0 | 0.0 | 0.0 |
| 06 | 0.0 | 2.655 | 2.92 | 490.673 | 0.0 | 0.833 | 0.0 | 0.0 | 0.0 | 0.0 | 1.273 | 0.0 | 0.0 | 0.0 | 0.0 | 0.808 |
| 07 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.803 | 2.685 | 0.0 | 0.0 | 0.0 | 4.193 | 0.0 | 0.0 | 0.0 | 491.481 | 0.0 |
| 08 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.803 | 0.0 | 1.179 | 0.0 | 494.256 | 2.924 | 0.0 | 0.0 | 0.0 | 0.0 |
| 09 | 1.179 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 17.91 | 464.168 | 0.884 | 12.096 | 0.0 | 2.924 | 0.0 | 0.0 |
| 10 | 5.002 | 2.966 | 9.942 | 1.179 | 11.084 | 84.289 | 321.105 | 50.614 | 0.0 | 0.0 | 0.884 | 0.0 | 1.176 | 1.149 | 7.769 | 2.002 |
| 11 | 0.0 | 5.334 | 11.928 | 0.0 | 1.833 | 3.057 | 82.705 | 0.809 | 23.351 | 52.395 | 243.518 | 20.436 | 3.021 | 1.963 | 43.976 | 4.835 |
| 12 | 1.227 | 22.198 | 2.797 | 1.984 | 0.84 | 44.134 | 5.399 | 12.376 | 39.197 | 316.381 | 13.233 | 13.316 | 0.0 | 23.127 | 0.934 | 2.021 |
| 13 | 4.349 | 21.685 | 13.993 | 1.238 | 30.178 | 150.237 | 145.173 | 80.253 | 1.816 | 8.935 | 9.671 | 1.939 | 1.06 | 6.978 | 17.534 | 4.125 |
| 14 | 6.656 | 4.931 | 20.389 | 5.428 | 45.132 | 43.209 | 74.58 | 24.914 | 18.271 | 88.356 | 72.04 | 7.704 | 6.487 | 33.245 | 33.924 | 13.898 |
| 15 | 15.348 | 31.415 | 17.899 | 11.883 | 14.089 | 97.854 | 33.052 | 24.745 | 29.788 | 100.167 | 45.488 | 25.491 | 2.674 | 31.23 | 9.743 | 8.296 |
| 16 | 7.895 | 15.088 | 27.856 | 11.059 | 38.231 | 131.292 | 72.418 | 18.726 | 13.012 | 30.539 | 48.584 | 14.048 | 7.802 | 18.891 | 32.807 | 10.917 |
| 17 | 11.31 | 7.873 | 36.442 | 11.315 | 76.387 | 23.006 | 64.924 | 31.491 | 19.419 | 61.658 | 81.916 | 18.672 | 6.774 | 16.512 | 18.121 | 13.342 |
| 18 | 9.112 | 25.199 | 58.096 | 21.484 | 8.737 | 39.72 | 26.741 | 33.85 | 28.521 | 86.68 | 61.343 | 24.859 | 5.895 | 21.492 | 28.455 | 18.979 |
| 19 | 9.947 | 8.38 | 29.031 | 4.907 | 35.727 | 41.515 | 60.824 | 35.025 | 26.379 | 52.966 | 69.904 | 25.386 | 9.5 | 25.65 | 48.223 | 15.799 |
| 20 | 15.43 | 10.692 | 46.355 | 9.075 | 17.298 | 27.361 | 55.602 | 28.249 | 28.458 | 55.276 | 100.606 | 23.642 | 9.913 | 25.08 | 31.31 | 14.815 |
| 21 | 15.294 | 14.222 | 25.912 | 15.67 | 19.993 | 36.02 | 27.099 | 35.298 | 52.461 | 78.835 | 67.993 | 34.583 | 6.095 | 39.753 | 16.975 | 12.96 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.953 | 0.41 | -0.52 | -1.601 | 0.35 | 1.313 | -1.555 | -0.212 | -0.331 | 0.612 | 0.023 | -1.123 | -0.492 | 0.706 | -1.273 | -1.119 |
| 02 | 0.969 | -3.244 | -0.802 | -4.399 | 2.125 | -2.222 | -0.633 | -2.519 | 0.569 | -2.658 | -1.58 | -2.734 | 0.181 | -2.244 | -1.062 | -4.399 |
| 03 | 2.53 | -1.278 | -0.837 | -0.346 | -4.399 | -2.632 | -2.644 | -1.783 | -1.031 | -0.741 | -1.249 | -0.888 | -3.241 | -2.519 | -4.399 | -3.267 |
| 04 | 1.055 | 0.855 | 1.96 | -0.448 | -0.859 | -1.925 | -1.327 | -2.618 | -2.303 | -0.954 | -2.083 | -1.644 | -1.5 | -0.084 | -2.261 | -2.549 |
| 05 | 1.276 | -4.399 | -4.399 | -4.399 | 1.315 | -4.399 | -2.945 | -4.399 | 2.016 | -3.253 | -4.399 | -3.273 | -0.055 | -4.399 | -4.399 | -4.399 |
| 06 | -4.399 | -2.34 | -2.256 | 2.744 | -4.399 | -3.253 | -4.399 | -4.399 | -4.399 | -4.399 | -2.945 | -4.399 | -4.399 | -4.399 | -4.399 | -3.273 |
| 07 | -4.399 | -4.399 | -4.399 | -4.399 | -4.399 | -3.277 | -2.33 | -4.399 | -4.399 | -4.399 | -1.931 | -4.399 | -4.399 | -4.399 | 2.746 | -4.399 |
| 08 | -4.399 | -4.399 | -4.399 | -4.399 | -4.399 | -4.399 | -3.277 | -4.399 | -3.003 | -4.399 | 2.751 | -2.255 | -4.399 | -4.399 | -4.399 | -4.399 |
| 09 | -3.003 | -4.399 | -4.399 | -4.399 | -4.399 | -4.399 | -4.399 | -4.399 | -0.546 | 2.688 | -3.212 | -0.928 | -4.399 | -2.255 | -4.399 | -4.399 |
| 10 | -1.768 | -2.242 | -1.118 | -3.003 | -1.013 | 0.986 | 2.32 | 0.479 | -4.399 | -4.399 | -3.212 | -4.399 | -3.005 | -3.023 | -1.354 | -2.581 |
| 11 | -4.399 | -1.708 | -0.942 | -4.399 | -2.655 | -2.216 | 0.967 | -3.273 | -0.286 | 0.513 | 2.044 | -0.417 | -2.226 | -2.598 | 0.34 | -1.8 |
| 12 | -2.973 | -0.335 | -2.294 | -2.589 | -3.247 | 0.343 | -1.697 | -0.906 | 0.226 | 2.305 | -0.841 | -0.835 | -4.399 | -0.295 | -3.174 | -2.574 |
| 13 | -1.897 | -0.358 | -0.787 | -2.967 | -0.033 | 1.562 | 1.528 | 0.937 | -2.662 | -1.22 | -1.144 | -2.608 | -3.082 | -1.456 | -0.567 | -1.946 |
| 14 | -1.501 | -1.781 | -0.419 | -1.692 | 0.365 | 0.322 | 0.864 | -0.222 | -0.526 | 1.033 | 0.83 | -1.362 | -1.525 | 0.063 | 0.083 | -0.793 |
| 15 | -0.697 | 0.007 | -0.546 | -0.945 | -0.78 | 1.135 | 0.057 | -0.229 | -0.046 | 1.158 | 0.373 | -0.199 | -2.334 | 0.001 | -1.137 | -1.291 |
| 16 | -1.338 | -0.713 | -0.112 | -1.015 | 0.201 | 1.428 | 0.835 | -0.502 | -0.857 | -0.021 | 0.439 | -0.783 | -1.35 | -0.494 | 0.05 | -1.027 |
| 17 | -0.993 | -1.341 | 0.154 | -0.993 | 0.888 | -0.3 | 0.726 | 0.009 | -0.467 | 0.675 | 0.958 | -0.505 | -1.484 | -0.625 | -0.534 | -0.833 |
| 18 | -1.201 | -0.211 | 0.616 | -0.367 | -1.242 | 0.239 | -0.152 | 0.081 | -0.089 | 1.014 | 0.67 | -0.224 | -1.615 | -0.367 | -0.091 | -0.489 |
| 19 | -1.117 | -1.282 | -0.071 | -1.786 | 0.134 | 0.283 | 0.662 | 0.114 | -0.166 | 0.524 | 0.8 | -0.203 | -1.161 | -0.193 | 0.431 | -0.668 |
| 20 | -0.692 | -1.048 | 0.392 | -1.205 | -0.58 | -0.13 | 0.572 | -0.098 | -0.091 | 0.567 | 1.162 | -0.273 | -1.12 | -0.215 | 0.004 | -0.731 |
| 21 | -0.7 | -0.771 | -0.183 | -0.676 | -0.438 | 0.142 | -0.139 | 0.122 | 0.515 | 0.92 | 0.772 | 0.102 | -1.583 | 0.24 | -0.598 | -0.861 |