Model info
| Transcription factor | USF2 (GeneCards) | ||||||||
| Model | USF2_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 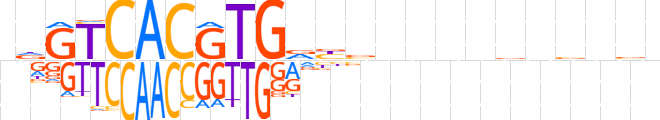 | ||||||||
| LOGO (reverse complement) | 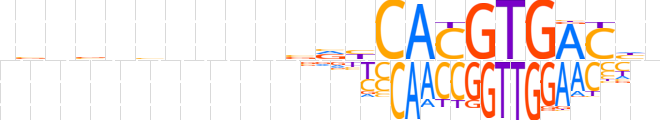 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv10 | ||||||||
| Model length | 23 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | ndGTCACGTGvbbnvnnbnvnbb | ||||||||
| Best auROC (human) | 0.998 | ||||||||
| Best auROC (mouse) | 0.999 | ||||||||
| Peak sets in benchmark (human) | 23 | ||||||||
| Peak sets in benchmark (mouse) | 10 | ||||||||
| Aligned words | 517 | ||||||||
| TF family | bHLH-ZIP factors {1.2.6} | ||||||||
| TF subfamily | USF factors {1.2.6.2} | ||||||||
| HGNC | HGNC:12594 | ||||||||
| EntrezGene | GeneID:7392 (SSTAR profile) | ||||||||
| UniProt ID | USF2_HUMAN | ||||||||
| UniProt AC | Q15853 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | USF2 expression | ||||||||
| ReMap ChIP-seq dataset list | USF2 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 20.032 | 7.901 | 71.456 | 14.163 | 36.198 | 20.403 | 39.671 | 32.523 | 52.947 | 11.473 | 71.279 | 22.146 | 15.084 | 5.683 | 74.101 | 4.94 |
| 02 | 13.499 | 1.108 | 109.653 | 0.0 | 9.217 | 0.0 | 36.243 | 0.0 | 51.274 | 5.985 | 199.248 | 0.0 | 5.564 | 0.972 | 67.236 | 0.0 |
| 03 | 3.565 | 9.553 | 3.252 | 63.184 | 0.0 | 2.964 | 0.0 | 5.101 | 10.899 | 15.852 | 11.069 | 374.559 | 0.0 | 0.0 | 0.0 | 0.0 |
| 04 | 0.0 | 14.465 | 0.0 | 0.0 | 0.0 | 28.369 | 0.0 | 0.0 | 0.0 | 14.322 | 0.0 | 0.0 | 0.0 | 442.844 | 0.0 | 0.0 |
| 05 | 0.0 | 0.0 | 0.0 | 0.0 | 497.627 | 0.0 | 0.0 | 2.373 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 06 | 0.0 | 489.802 | 0.794 | 7.031 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.373 | 0.0 | 0.0 |
| 07 | 0.0 | 0.0 | 0.0 | 0.0 | 83.357 | 6.48 | 402.338 | 0.0 | 0.0 | 0.794 | 0.0 | 0.0 | 0.0 | 0.0 | 7.031 | 0.0 |
| 08 | 0.0 | 0.0 | 0.0 | 83.357 | 0.0 | 0.0 | 0.0 | 7.274 | 1.828 | 3.636 | 2.916 | 400.989 | 0.0 | 0.0 | 0.0 | 0.0 |
| 09 | 0.0 | 0.0 | 1.828 | 0.0 | 0.0 | 0.0 | 3.636 | 0.0 | 0.0 | 0.0 | 2.916 | 0.0 | 0.0 | 0.0 | 491.62 | 0.0 |
| 10 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 239.956 | 44.129 | 173.704 | 42.211 | 0.0 | 0.0 | 0.0 | 0.0 |
| 11 | 3.624 | 136.374 | 22.723 | 77.236 | 1.928 | 12.894 | 9.633 | 19.674 | 10.887 | 51.799 | 49.342 | 61.676 | 1.153 | 22.38 | 10.116 | 8.562 |
| 12 | 2.835 | 7.089 | 7.668 | 0.0 | 32.173 | 98.286 | 41.976 | 51.013 | 10.226 | 44.826 | 23.403 | 13.359 | 12.819 | 65.74 | 71.282 | 17.305 |
| 13 | 5.357 | 17.539 | 26.808 | 8.349 | 67.377 | 51.119 | 52.635 | 44.81 | 20.418 | 64.845 | 41.305 | 17.761 | 10.941 | 22.509 | 32.166 | 16.06 |
| 14 | 20.127 | 33.944 | 35.579 | 14.444 | 40.265 | 41.174 | 38.641 | 35.931 | 25.92 | 60.541 | 58.284 | 8.17 | 9.62 | 35.046 | 30.549 | 11.766 |
| 15 | 25.22 | 12.187 | 48.833 | 9.693 | 34.157 | 62.287 | 36.329 | 37.931 | 36.836 | 55.49 | 54.792 | 15.934 | 8.516 | 29.899 | 22.262 | 9.634 |
| 16 | 19.247 | 26.3 | 48.368 | 10.813 | 47.085 | 44.395 | 40.55 | 27.833 | 39.959 | 61.115 | 35.21 | 25.931 | 6.557 | 21.106 | 37.184 | 8.345 |
| 17 | 16.785 | 25.915 | 54.645 | 15.503 | 28.798 | 48.548 | 37.657 | 37.913 | 25.322 | 55.658 | 58.996 | 21.337 | 2.923 | 18.251 | 42.383 | 9.365 |
| 18 | 10.699 | 15.291 | 37.248 | 10.591 | 30.219 | 28.705 | 38.21 | 51.239 | 32.041 | 47.838 | 76.857 | 36.944 | 7.389 | 26.813 | 28.49 | 21.426 |
| 19 | 16.25 | 23.085 | 28.928 | 12.085 | 25.735 | 34.901 | 36.117 | 21.893 | 25.119 | 59.449 | 75.675 | 20.562 | 10.997 | 37.115 | 54.02 | 18.068 |
| 20 | 22.914 | 15.734 | 31.949 | 7.505 | 41.397 | 51.924 | 29.67 | 31.559 | 31.959 | 60.744 | 79.443 | 22.594 | 7.363 | 24.11 | 33.382 | 7.754 |
| 21 | 10.721 | 35.342 | 45.304 | 12.265 | 28.903 | 57.531 | 36.076 | 30.002 | 25.957 | 68.855 | 53.964 | 25.667 | 9.97 | 20.934 | 28.998 | 9.51 |
| 22 | 9.364 | 31.034 | 22.457 | 12.697 | 34.894 | 67.784 | 34.385 | 45.6 | 16.875 | 70.429 | 53.963 | 23.075 | 5.228 | 22.162 | 32.491 | 17.563 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.438 | -1.339 | 0.82 | -0.777 | 0.145 | -0.42 | 0.236 | 0.039 | 0.522 | -0.981 | 0.818 | -0.339 | -0.715 | -1.651 | 0.856 | -1.781 |
| 02 | -0.823 | -3.051 | 1.246 | -4.4 | -1.192 | -4.4 | 0.147 | -4.4 | 0.49 | -1.602 | 1.842 | -4.4 | -1.671 | -3.147 | 0.76 | -4.4 |
| 03 | -2.08 | -1.158 | -2.162 | 0.698 | -4.4 | -2.245 | -4.4 | -1.752 | -1.031 | -0.667 | -1.016 | 2.472 | -4.4 | -4.4 | -4.4 | -4.4 |
| 04 | -4.4 | -0.756 | -4.4 | -4.4 | -4.4 | -0.095 | -4.4 | -4.4 | -4.4 | -0.766 | -4.4 | -4.4 | -4.4 | 2.64 | -4.4 | -4.4 |
| 05 | -4.4 | -4.4 | -4.4 | -4.4 | 2.756 | -4.4 | -4.4 | -2.439 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 |
| 06 | -4.4 | 2.74 | -3.286 | -1.45 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -2.439 | -4.4 | -4.4 |
| 07 | -4.4 | -4.4 | -4.4 | -4.4 | 0.973 | -1.528 | 2.544 | -4.4 | -4.4 | -3.286 | -4.4 | -4.4 | -4.4 | -4.4 | -1.45 | -4.4 |
| 08 | -4.4 | -4.4 | -4.4 | 0.973 | -4.4 | -4.4 | -4.4 | -1.418 | -2.659 | -2.062 | -2.259 | 2.541 | -4.4 | -4.4 | -4.4 | -4.4 |
| 09 | -4.4 | -4.4 | -2.659 | -4.4 | -4.4 | -4.4 | -2.062 | -4.4 | -4.4 | -4.4 | -2.259 | -4.4 | -4.4 | -4.4 | 2.744 | -4.4 |
| 10 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | 2.028 | 0.342 | 1.705 | 0.297 | -4.4 | -4.4 | -4.4 | -4.4 |
| 11 | -2.065 | 1.464 | -0.314 | 0.898 | -2.614 | -0.868 | -1.15 | -0.456 | -1.032 | 0.5 | 0.452 | 0.674 | -3.022 | -0.329 | -1.103 | -1.263 |
| 12 | -2.284 | -1.442 | -1.368 | -4.4 | 0.029 | 1.137 | 0.292 | 0.485 | -1.092 | 0.357 | -0.285 | -0.834 | -0.874 | 0.737 | 0.818 | -0.581 |
| 13 | -1.706 | -0.568 | -0.151 | -1.287 | 0.762 | 0.487 | 0.516 | 0.357 | -0.419 | 0.724 | 0.276 | -0.556 | -1.027 | -0.323 | 0.029 | -0.654 |
| 14 | -0.433 | 0.082 | 0.128 | -0.758 | 0.251 | 0.273 | 0.21 | 0.138 | -0.184 | 0.655 | 0.618 | -1.307 | -1.151 | 0.113 | -0.022 | -0.957 |
| 15 | -0.211 | -0.923 | 0.442 | -1.144 | 0.088 | 0.684 | 0.149 | 0.192 | 0.163 | 0.569 | 0.556 | -0.662 | -1.268 | -0.044 | -0.334 | -1.15 |
| 16 | -0.477 | -0.17 | 0.432 | -1.038 | 0.406 | 0.347 | 0.258 | -0.114 | 0.243 | 0.665 | 0.118 | -0.184 | -1.516 | -0.387 | 0.172 | -1.287 |
| 17 | -0.611 | -0.185 | 0.554 | -0.689 | -0.081 | 0.436 | 0.184 | 0.191 | -0.207 | 0.572 | 0.63 | -0.376 | -2.257 | -0.529 | 0.301 | -1.177 |
| 18 | -1.049 | -0.702 | 0.174 | -1.058 | -0.033 | -0.084 | 0.199 | 0.49 | 0.025 | 0.422 | 0.893 | 0.165 | -1.403 | -0.151 | -0.091 | -0.372 |
| 19 | -0.643 | -0.298 | -0.076 | -0.931 | -0.192 | 0.109 | 0.143 | -0.351 | -0.215 | 0.637 | 0.877 | -0.412 | -1.022 | 0.17 | 0.542 | -0.539 |
| 20 | -0.306 | -0.674 | 0.022 | -1.388 | 0.278 | 0.503 | -0.051 | 0.01 | 0.022 | 0.659 | 0.926 | -0.32 | -1.407 | -0.256 | 0.065 | -1.357 |
| 21 | -1.047 | 0.122 | 0.368 | -0.916 | -0.077 | 0.605 | 0.142 | -0.04 | -0.183 | 0.783 | 0.541 | -0.194 | -1.117 | -0.395 | -0.074 | -1.162 |
| 22 | -1.177 | -0.007 | -0.326 | -0.883 | 0.109 | 0.768 | 0.094 | 0.374 | -0.606 | 0.806 | 0.541 | -0.299 | -1.729 | -0.339 | 0.038 | -0.567 |