Model info
| Transcription factor | VDR (GeneCards) | ||||||||
| Model | VDR_HUMAN.H11MO.0.A | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 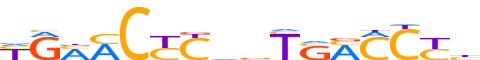 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 16 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | vRGKKSAbhRRGKKYR | ||||||||
| Best auROC (human) | 0.943 | ||||||||
| Best auROC (mouse) | 0.932 | ||||||||
| Peak sets in benchmark (human) | 48 | ||||||||
| Peak sets in benchmark (mouse) | 30 | ||||||||
| Aligned words | 500 | ||||||||
| TF family | Thyroid hormone receptor-related factors (NR1) {2.1.2} | ||||||||
| TF subfamily | Vitamin D receptor (NR1I) {2.1.2.4} | ||||||||
| HGNC | HGNC:12679 | ||||||||
| EntrezGene | GeneID:7421 (SSTAR profile) | ||||||||
| UniProt ID | VDR_HUMAN | ||||||||
| UniProt AC | P11473 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | VDR expression | ||||||||
| ReMap ChIP-seq dataset list | VDR datasets | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 139.0 | 67.0 | 230.0 | 64.0 |
| 02 | 233.0 | 14.0 | 246.0 | 7.0 |
| 03 | 59.0 | 4.0 | 422.0 | 15.0 |
| 04 | 26.0 | 4.0 | 381.0 | 89.0 |
| 05 | 5.0 | 62.0 | 93.0 | 340.0 |
| 06 | 25.0 | 345.0 | 79.0 | 51.0 |
| 07 | 406.0 | 7.0 | 54.0 | 33.0 |
| 08 | 68.0 | 169.0 | 174.0 | 89.0 |
| 09 | 104.0 | 110.0 | 77.0 | 209.0 |
| 10 | 72.0 | 30.0 | 386.0 | 12.0 |
| 11 | 237.0 | 9.0 | 248.0 | 6.0 |
| 12 | 4.0 | 2.0 | 488.0 | 6.0 |
| 13 | 8.0 | 4.0 | 184.0 | 304.0 |
| 14 | 39.0 | 40.0 | 98.0 | 323.0 |
| 15 | 4.0 | 384.0 | 30.0 | 82.0 |
| 16 | 328.0 | 42.0 | 79.0 | 51.0 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.105 | -0.613 | 0.604 | -0.658 |
| 02 | 0.617 | -2.096 | 0.671 | -2.694 |
| 03 | -0.737 | -3.126 | 1.208 | -2.034 |
| 04 | -1.525 | -3.126 | 1.106 | -0.335 |
| 05 | -2.961 | -0.689 | -0.291 | 0.993 |
| 06 | -1.561 | 1.007 | -0.452 | -0.879 |
| 07 | 1.17 | -2.694 | -0.823 | -1.298 |
| 08 | -0.599 | 0.298 | 0.327 | -0.335 |
| 09 | -0.181 | -0.126 | -0.477 | 0.509 |
| 10 | -0.543 | -1.389 | 1.119 | -2.234 |
| 11 | 0.634 | -2.484 | 0.679 | -2.819 |
| 12 | -3.126 | -3.573 | 1.353 | -2.819 |
| 13 | -2.584 | -3.126 | 0.383 | 0.881 |
| 14 | -1.138 | -1.114 | -0.24 | 0.942 |
| 15 | -3.126 | 1.114 | -1.389 | -0.415 |
| 16 | 0.957 | -1.067 | -0.452 | -0.879 |