Model info
| Transcription factor | VEZF1 (GeneCards) | ||||||||
| Model | VEZF1_HUMAN.H11DI.0.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 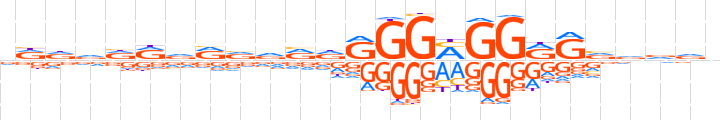 | ||||||||
| LOGO (reverse complement) | 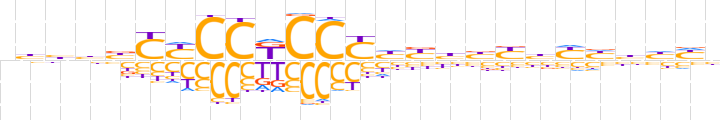 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 25 | ||||||||
| Quality | C | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | dddvdRvRvdRdRGGMGGRRvvddn | ||||||||
| Best auROC (human) | 0.907 | ||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | 5 | ||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 498 | ||||||||
| TF family | Factors with multiple dispersed zinc fingers {2.3.4} | ||||||||
| TF subfamily | MAZ-like factors {2.3.4.8} | ||||||||
| HGNC | HGNC:12949 | ||||||||
| EntrezGene | GeneID:7716 (SSTAR profile) | ||||||||
| UniProt ID | VEZF1_HUMAN | ||||||||
| UniProt AC | Q14119 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | VEZF1 expression | ||||||||
| ReMap ChIP-seq dataset list | VEZF1 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 16.0 | 11.0 | 84.0 | 9.0 | 12.0 | 7.0 | 36.0 | 28.0 | 47.0 | 15.0 | 115.0 | 27.0 | 13.0 | 12.0 | 54.0 | 12.0 |
| 02 | 33.0 | 2.0 | 37.0 | 16.0 | 3.0 | 8.0 | 18.0 | 16.0 | 76.0 | 29.0 | 165.0 | 19.0 | 4.0 | 11.0 | 45.0 | 16.0 |
| 03 | 56.0 | 6.0 | 45.0 | 9.0 | 17.0 | 14.0 | 18.0 | 1.0 | 86.0 | 49.0 | 100.0 | 30.0 | 12.0 | 3.0 | 38.0 | 14.0 |
| 04 | 44.0 | 7.0 | 100.0 | 20.0 | 14.0 | 12.0 | 31.0 | 15.0 | 23.0 | 27.0 | 128.0 | 23.0 | 10.0 | 7.0 | 28.0 | 9.0 |
| 05 | 33.0 | 4.0 | 44.0 | 10.0 | 15.0 | 0.0 | 32.0 | 6.0 | 47.0 | 18.0 | 185.0 | 37.0 | 13.0 | 1.0 | 41.0 | 12.0 |
| 06 | 32.0 | 11.0 | 51.0 | 14.0 | 6.0 | 8.0 | 6.0 | 3.0 | 83.0 | 50.0 | 138.0 | 31.0 | 9.0 | 11.0 | 36.0 | 9.0 |
| 07 | 45.0 | 3.0 | 73.0 | 9.0 | 21.0 | 13.0 | 34.0 | 12.0 | 49.0 | 28.0 | 142.0 | 12.0 | 16.0 | 2.0 | 37.0 | 2.0 |
| 08 | 40.0 | 4.0 | 74.0 | 13.0 | 20.0 | 7.0 | 14.0 | 5.0 | 56.0 | 50.0 | 160.0 | 20.0 | 7.0 | 3.0 | 19.0 | 6.0 |
| 09 | 48.0 | 9.0 | 56.0 | 10.0 | 22.0 | 5.0 | 29.0 | 8.0 | 103.0 | 31.0 | 106.0 | 27.0 | 9.0 | 7.0 | 19.0 | 9.0 |
| 10 | 80.0 | 20.0 | 81.0 | 1.0 | 14.0 | 7.0 | 25.0 | 6.0 | 61.0 | 33.0 | 109.0 | 7.0 | 12.0 | 5.0 | 33.0 | 4.0 |
| 11 | 55.0 | 13.0 | 76.0 | 23.0 | 16.0 | 8.0 | 29.0 | 12.0 | 79.0 | 13.0 | 130.0 | 26.0 | 3.0 | 0.0 | 13.0 | 2.0 |
| 12 | 45.0 | 5.0 | 99.0 | 4.0 | 12.0 | 2.0 | 19.0 | 1.0 | 64.0 | 5.0 | 175.0 | 4.0 | 4.0 | 4.0 | 48.0 | 7.0 |
| 13 | 4.0 | 0.0 | 118.0 | 3.0 | 2.0 | 0.0 | 11.0 | 3.0 | 13.0 | 3.0 | 305.0 | 20.0 | 3.0 | 0.0 | 12.0 | 1.0 |
| 14 | 2.0 | 0.0 | 20.0 | 0.0 | 0.0 | 1.0 | 2.0 | 0.0 | 1.0 | 15.0 | 415.0 | 15.0 | 0.0 | 2.0 | 24.0 | 1.0 |
| 15 | 0.0 | 1.0 | 0.0 | 2.0 | 13.0 | 4.0 | 0.0 | 1.0 | 282.0 | 109.0 | 1.0 | 69.0 | 6.0 | 5.0 | 0.0 | 5.0 |
| 16 | 39.0 | 1.0 | 258.0 | 3.0 | 10.0 | 1.0 | 105.0 | 3.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 76.0 | 0.0 |
| 17 | 2.0 | 1.0 | 46.0 | 1.0 | 0.0 | 0.0 | 2.0 | 0.0 | 22.0 | 6.0 | 406.0 | 6.0 | 0.0 | 0.0 | 6.0 | 0.0 |
| 18 | 6.0 | 2.0 | 12.0 | 4.0 | 3.0 | 0.0 | 3.0 | 1.0 | 142.0 | 7.0 | 269.0 | 42.0 | 3.0 | 0.0 | 3.0 | 1.0 |
| 19 | 53.0 | 6.0 | 92.0 | 3.0 | 2.0 | 0.0 | 7.0 | 0.0 | 59.0 | 2.0 | 218.0 | 8.0 | 11.0 | 0.0 | 36.0 | 1.0 |
| 20 | 51.0 | 12.0 | 56.0 | 6.0 | 1.0 | 3.0 | 3.0 | 1.0 | 86.0 | 82.0 | 166.0 | 19.0 | 1.0 | 0.0 | 11.0 | 0.0 |
| 21 | 48.0 | 18.0 | 63.0 | 10.0 | 20.0 | 30.0 | 32.0 | 15.0 | 76.0 | 51.0 | 85.0 | 24.0 | 11.0 | 4.0 | 8.0 | 3.0 |
| 22 | 58.0 | 17.0 | 50.0 | 30.0 | 44.0 | 10.0 | 29.0 | 20.0 | 92.0 | 14.0 | 46.0 | 36.0 | 5.0 | 5.0 | 31.0 | 11.0 |
| 23 | 61.0 | 21.0 | 85.0 | 32.0 | 12.0 | 2.0 | 20.0 | 12.0 | 46.0 | 21.0 | 73.0 | 16.0 | 19.0 | 13.0 | 59.0 | 6.0 |
| 24 | 43.0 | 28.0 | 49.0 | 18.0 | 17.0 | 13.0 | 17.0 | 10.0 | 45.0 | 63.0 | 86.0 | 43.0 | 11.0 | 16.0 | 28.0 | 11.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.654 | -1.018 | 0.985 | -1.211 | -0.934 | -1.451 | 0.144 | -0.104 | 0.408 | -0.717 | 1.298 | -0.14 | -0.856 | -0.934 | 0.546 | -0.934 |
| 02 | 0.058 | -2.58 | 0.171 | -0.654 | -2.23 | -1.324 | -0.539 | -0.654 | 0.885 | -0.07 | 1.658 | -0.486 | -1.971 | -1.018 | 0.365 | -0.654 |
| 03 | 0.582 | -1.596 | 0.365 | -1.211 | -0.595 | -0.784 | -0.539 | -3.122 | 1.008 | 0.449 | 1.159 | -0.036 | -0.934 | -2.23 | 0.197 | -0.784 |
| 04 | 0.343 | -1.451 | 1.159 | -0.435 | -0.784 | -0.934 | -0.004 | -0.717 | -0.298 | -0.14 | 1.405 | -0.298 | -1.11 | -1.451 | -0.104 | -1.211 |
| 05 | 0.058 | -1.971 | 0.343 | -1.11 | -0.717 | -4.397 | 0.027 | -1.596 | 0.408 | -0.539 | 1.772 | 0.171 | -0.856 | -3.122 | 0.273 | -0.934 |
| 06 | 0.027 | -1.018 | 0.489 | -0.784 | -1.596 | -1.324 | -1.596 | -2.23 | 0.973 | 0.469 | 1.48 | -0.004 | -1.211 | -1.018 | 0.144 | -1.211 |
| 07 | 0.365 | -2.23 | 0.845 | -1.211 | -0.388 | -0.856 | 0.087 | -0.934 | 0.449 | -0.104 | 1.508 | -0.934 | -0.654 | -2.58 | 0.171 | -2.58 |
| 08 | 0.248 | -1.971 | 0.859 | -0.856 | -0.435 | -1.451 | -0.784 | -1.766 | 0.582 | 0.469 | 1.627 | -0.435 | -1.451 | -2.23 | -0.486 | -1.596 |
| 09 | 0.429 | -1.211 | 0.582 | -1.11 | -0.342 | -1.766 | -0.07 | -1.324 | 1.188 | -0.004 | 1.217 | -0.14 | -1.211 | -1.451 | -0.486 | -1.211 |
| 10 | 0.936 | -0.435 | 0.949 | -3.122 | -0.784 | -1.451 | -0.216 | -1.596 | 0.667 | 0.058 | 1.244 | -1.451 | -0.934 | -1.766 | 0.058 | -1.971 |
| 11 | 0.564 | -0.856 | 0.885 | -0.298 | -0.654 | -1.324 | -0.07 | -0.934 | 0.924 | -0.856 | 1.42 | -0.177 | -2.23 | -4.397 | -0.856 | -2.58 |
| 12 | 0.365 | -1.766 | 1.149 | -1.971 | -0.934 | -2.58 | -0.486 | -3.122 | 0.715 | -1.766 | 1.717 | -1.971 | -1.971 | -1.971 | 0.429 | -1.451 |
| 13 | -1.971 | -4.397 | 1.324 | -2.23 | -2.58 | -4.397 | -1.018 | -2.23 | -0.856 | -2.23 | 2.271 | -0.435 | -2.23 | -4.397 | -0.934 | -3.122 |
| 14 | -2.58 | -4.397 | -0.435 | -4.397 | -4.397 | -3.122 | -2.58 | -4.397 | -3.122 | -0.717 | 2.579 | -0.717 | -4.397 | -2.58 | -0.256 | -3.122 |
| 15 | -4.397 | -3.122 | -4.397 | -2.58 | -0.856 | -1.971 | -4.397 | -3.122 | 2.193 | 1.244 | -3.122 | 0.789 | -1.596 | -1.766 | -4.397 | -1.766 |
| 16 | 0.223 | -3.122 | 2.104 | -2.23 | -1.11 | -3.122 | 1.207 | -2.23 | -4.397 | -4.397 | -3.122 | -4.397 | -3.122 | -4.397 | 0.885 | -4.397 |
| 17 | -2.58 | -3.122 | 0.387 | -3.122 | -4.397 | -4.397 | -2.58 | -4.397 | -0.342 | -1.596 | 2.557 | -1.596 | -4.397 | -4.397 | -1.596 | -4.397 |
| 18 | -1.596 | -2.58 | -0.934 | -1.971 | -2.23 | -4.397 | -2.23 | -3.122 | 1.508 | -1.451 | 2.146 | 0.296 | -2.23 | -4.397 | -2.23 | -3.122 |
| 19 | 0.527 | -1.596 | 1.076 | -2.23 | -2.58 | -4.397 | -1.451 | -4.397 | 0.634 | -2.58 | 1.936 | -1.324 | -1.018 | -4.397 | 0.144 | -3.122 |
| 20 | 0.489 | -0.934 | 0.582 | -1.596 | -3.122 | -2.23 | -2.23 | -3.122 | 1.008 | 0.961 | 1.664 | -0.486 | -3.122 | -4.397 | -1.018 | -4.397 |
| 21 | 0.429 | -0.539 | 0.699 | -1.11 | -0.435 | -0.036 | 0.027 | -0.717 | 0.885 | 0.489 | 0.997 | -0.256 | -1.018 | -1.971 | -1.324 | -2.23 |
| 22 | 0.617 | -0.595 | 0.469 | -0.036 | 0.343 | -1.11 | -0.07 | -0.435 | 1.076 | -0.784 | 0.387 | 0.144 | -1.766 | -1.766 | -0.004 | -1.018 |
| 23 | 0.667 | -0.388 | 0.997 | 0.027 | -0.934 | -2.58 | -0.435 | -0.934 | 0.387 | -0.388 | 0.845 | -0.654 | -0.486 | -0.856 | 0.634 | -1.596 |
| 24 | 0.32 | -0.104 | 0.449 | -0.539 | -0.595 | -0.856 | -0.595 | -1.11 | 0.365 | 0.699 | 1.008 | 0.32 | -1.018 | -0.654 | -0.104 | -1.018 |