Model info
| Transcription factor | Wt1 | ||||||||
| Model | WT1_MOUSE.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 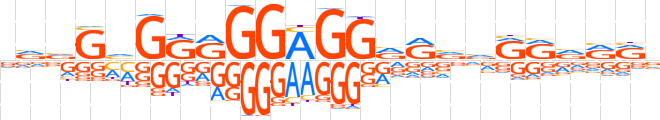 | ||||||||
| LOGO (reverse complement) | 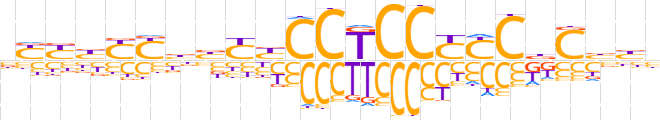 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 23 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | vvvGhGRRGGAGGRRvvRRvRRv | ||||||||
| Best auROC (human) | |||||||||
| Best auROC (mouse) | 0.99 | ||||||||
| Peak sets in benchmark (human) | |||||||||
| Peak sets in benchmark (mouse) | 5 | ||||||||
| Aligned words | 481 | ||||||||
| TF family | More than 3 adjacent zinc finger factors {2.3.3} | ||||||||
| TF subfamily | unclassified {2.3.3.0} | ||||||||
| MGI | MGI:98968 | ||||||||
| EntrezGene | |||||||||
| UniProt ID | WT1_MOUSE | ||||||||
| UniProt AC | P22561 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Wt1 expression | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 31.0 | 13.0 | 53.0 | 7.0 | 24.0 | 10.0 | 38.0 | 16.0 | 73.0 | 34.0 | 118.0 | 4.0 | 5.0 | 4.0 | 21.0 | 6.0 |
| 02 | 23.0 | 9.0 | 90.0 | 11.0 | 16.0 | 11.0 | 26.0 | 8.0 | 41.0 | 55.0 | 105.0 | 29.0 | 6.0 | 5.0 | 16.0 | 6.0 |
| 03 | 4.0 | 5.0 | 76.0 | 1.0 | 1.0 | 13.0 | 63.0 | 3.0 | 10.0 | 18.0 | 204.0 | 5.0 | 3.0 | 5.0 | 42.0 | 4.0 |
| 04 | 4.0 | 4.0 | 4.0 | 6.0 | 10.0 | 16.0 | 2.0 | 13.0 | 142.0 | 165.0 | 33.0 | 45.0 | 4.0 | 1.0 | 7.0 | 1.0 |
| 05 | 5.0 | 1.0 | 153.0 | 1.0 | 7.0 | 2.0 | 173.0 | 4.0 | 7.0 | 1.0 | 37.0 | 1.0 | 1.0 | 1.0 | 61.0 | 2.0 |
| 06 | 6.0 | 2.0 | 10.0 | 2.0 | 1.0 | 1.0 | 2.0 | 1.0 | 47.0 | 7.0 | 324.0 | 46.0 | 0.0 | 0.0 | 6.0 | 2.0 |
| 07 | 10.0 | 1.0 | 43.0 | 0.0 | 4.0 | 0.0 | 6.0 | 0.0 | 135.0 | 13.0 | 194.0 | 0.0 | 3.0 | 2.0 | 44.0 | 2.0 |
| 08 | 6.0 | 0.0 | 145.0 | 1.0 | 3.0 | 0.0 | 13.0 | 0.0 | 1.0 | 1.0 | 285.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 |
| 09 | 1.0 | 0.0 | 9.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 2.0 | 3.0 | 439.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 |
| 10 | 3.0 | 0.0 | 0.0 | 0.0 | 2.0 | 1.0 | 0.0 | 0.0 | 359.0 | 65.0 | 0.0 | 26.0 | 1.0 | 0.0 | 0.0 | 0.0 |
| 11 | 4.0 | 1.0 | 355.0 | 5.0 | 6.0 | 1.0 | 58.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 26.0 | 0.0 |
| 12 | 0.0 | 0.0 | 10.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 15.0 | 6.0 | 400.0 | 18.0 | 0.0 | 0.0 | 6.0 | 0.0 |
| 13 | 7.0 | 2.0 | 5.0 | 1.0 | 1.0 | 1.0 | 4.0 | 0.0 | 165.0 | 44.0 | 195.0 | 14.0 | 5.0 | 4.0 | 8.0 | 1.0 |
| 14 | 44.0 | 5.0 | 126.0 | 3.0 | 10.0 | 5.0 | 31.0 | 5.0 | 80.0 | 8.0 | 121.0 | 3.0 | 5.0 | 0.0 | 9.0 | 2.0 |
| 15 | 32.0 | 6.0 | 97.0 | 4.0 | 3.0 | 5.0 | 6.0 | 4.0 | 71.0 | 76.0 | 125.0 | 15.0 | 2.0 | 0.0 | 11.0 | 0.0 |
| 16 | 23.0 | 24.0 | 48.0 | 13.0 | 22.0 | 13.0 | 42.0 | 10.0 | 98.0 | 57.0 | 70.0 | 14.0 | 2.0 | 4.0 | 15.0 | 2.0 |
| 17 | 17.0 | 7.0 | 114.0 | 7.0 | 10.0 | 12.0 | 72.0 | 4.0 | 36.0 | 13.0 | 122.0 | 4.0 | 2.0 | 1.0 | 30.0 | 6.0 |
| 18 | 9.0 | 5.0 | 48.0 | 3.0 | 12.0 | 2.0 | 16.0 | 3.0 | 51.0 | 29.0 | 244.0 | 14.0 | 4.0 | 3.0 | 13.0 | 1.0 |
| 19 | 23.0 | 4.0 | 48.0 | 1.0 | 8.0 | 3.0 | 23.0 | 5.0 | 123.0 | 45.0 | 135.0 | 18.0 | 5.0 | 3.0 | 13.0 | 0.0 |
| 20 | 45.0 | 13.0 | 101.0 | 0.0 | 8.0 | 4.0 | 37.0 | 6.0 | 73.0 | 37.0 | 108.0 | 1.0 | 5.0 | 1.0 | 15.0 | 3.0 |
| 21 | 30.0 | 9.0 | 82.0 | 10.0 | 11.0 | 8.0 | 30.0 | 6.0 | 42.0 | 25.0 | 179.0 | 15.0 | 0.0 | 2.0 | 6.0 | 2.0 |
| 22 | 18.0 | 12.0 | 42.0 | 11.0 | 8.0 | 5.0 | 18.0 | 13.0 | 57.0 | 82.0 | 125.0 | 33.0 | 6.0 | 4.0 | 14.0 | 9.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.081 | -0.771 | 0.612 | -1.366 | -0.172 | -1.025 | 0.282 | -0.569 | 0.93 | 0.172 | 1.409 | -1.888 | -1.682 | -1.888 | -0.303 | -1.512 |
| 02 | -0.213 | -1.127 | 1.139 | -0.933 | -0.569 | -0.933 | -0.093 | -1.239 | 0.357 | 0.649 | 1.292 | 0.015 | -1.512 | -1.682 | -0.569 | -1.512 |
| 03 | -1.888 | -1.682 | 0.97 | -3.041 | -3.041 | -0.771 | 0.784 | -2.147 | -1.025 | -0.454 | 1.955 | -1.682 | -2.147 | -1.682 | 0.381 | -1.888 |
| 04 | -1.888 | -1.888 | -1.888 | -1.512 | -1.025 | -0.569 | -2.497 | -0.771 | 1.593 | 1.743 | 0.143 | 0.45 | -1.888 | -3.041 | -1.366 | -3.041 |
| 05 | -1.682 | -3.041 | 1.668 | -3.041 | -1.366 | -2.497 | 1.79 | -1.888 | -1.366 | -3.041 | 0.256 | -3.041 | -3.041 | -3.041 | 0.752 | -2.497 |
| 06 | -1.512 | -2.497 | -1.025 | -2.497 | -3.041 | -3.041 | -2.497 | -3.041 | 0.493 | -1.366 | 2.417 | 0.472 | -4.326 | -4.326 | -1.512 | -2.497 |
| 07 | -1.025 | -3.041 | 0.405 | -4.326 | -1.888 | -4.326 | -1.512 | -4.326 | 1.543 | -0.771 | 1.904 | -4.326 | -2.147 | -2.497 | 0.427 | -2.497 |
| 08 | -1.512 | -4.326 | 1.614 | -3.041 | -2.147 | -4.326 | -0.771 | -4.326 | -3.041 | -3.041 | 2.288 | -4.326 | -4.326 | -4.326 | -2.497 | -4.326 |
| 09 | -3.041 | -4.326 | -1.127 | -4.326 | -4.326 | -4.326 | -3.041 | -4.326 | -2.497 | -2.147 | 2.72 | -3.041 | -4.326 | -4.326 | -3.041 | -4.326 |
| 10 | -2.147 | -4.326 | -4.326 | -4.326 | -2.497 | -3.041 | -4.326 | -4.326 | 2.519 | 0.815 | -4.326 | -0.093 | -3.041 | -4.326 | -4.326 | -4.326 |
| 11 | -1.888 | -3.041 | 2.508 | -1.682 | -1.512 | -3.041 | 0.702 | -3.041 | -4.326 | -4.326 | -4.326 | -4.326 | -4.326 | -4.326 | -0.093 | -4.326 |
| 12 | -4.326 | -4.326 | -1.025 | -4.326 | -4.326 | -4.326 | -2.497 | -4.326 | -0.632 | -1.512 | 2.627 | -0.454 | -4.326 | -4.326 | -1.512 | -4.326 |
| 13 | -1.366 | -2.497 | -1.682 | -3.041 | -3.041 | -3.041 | -1.888 | -4.326 | 1.743 | 0.427 | 1.91 | -0.699 | -1.682 | -1.888 | -1.239 | -3.041 |
| 14 | 0.427 | -1.682 | 1.474 | -2.147 | -1.025 | -1.682 | 0.081 | -1.682 | 1.021 | -1.239 | 1.434 | -2.147 | -1.682 | -4.326 | -1.127 | -2.497 |
| 15 | 0.112 | -1.512 | 1.213 | -1.888 | -2.147 | -1.682 | -1.512 | -1.888 | 0.903 | 0.97 | 1.466 | -0.632 | -2.497 | -4.326 | -0.933 | -4.326 |
| 16 | -0.213 | -0.172 | 0.514 | -0.771 | -0.257 | -0.771 | 0.381 | -1.025 | 1.223 | 0.684 | 0.889 | -0.699 | -2.497 | -1.888 | -0.632 | -2.497 |
| 17 | -0.51 | -1.366 | 1.374 | -1.366 | -1.025 | -0.849 | 0.917 | -1.888 | 0.229 | -0.771 | 1.442 | -1.888 | -2.497 | -3.041 | 0.048 | -1.512 |
| 18 | -1.127 | -1.682 | 0.514 | -2.147 | -0.849 | -2.497 | -0.569 | -2.147 | 0.574 | 0.015 | 2.133 | -0.699 | -1.888 | -2.147 | -0.771 | -3.041 |
| 19 | -0.213 | -1.888 | 0.514 | -3.041 | -1.239 | -2.147 | -0.213 | -1.682 | 1.45 | 0.45 | 1.543 | -0.454 | -1.682 | -2.147 | -0.771 | -4.326 |
| 20 | 0.45 | -0.771 | 1.253 | -4.326 | -1.239 | -1.888 | 0.256 | -1.512 | 0.93 | 0.256 | 1.32 | -3.041 | -1.682 | -3.041 | -0.632 | -2.147 |
| 21 | 0.048 | -1.127 | 1.046 | -1.025 | -0.933 | -1.239 | 0.048 | -1.512 | 0.381 | -0.131 | 1.824 | -0.632 | -4.326 | -2.497 | -1.512 | -2.497 |
| 22 | -0.454 | -0.849 | 0.381 | -0.933 | -1.239 | -1.682 | -0.454 | -0.771 | 0.684 | 1.046 | 1.466 | 0.143 | -1.512 | -1.888 | -0.699 | -1.127 |