Model info
| Transcription factor | ZBTB18 (GeneCards) | ||||||||
| Model | ZBT18_HUMAN.H11DI.0.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 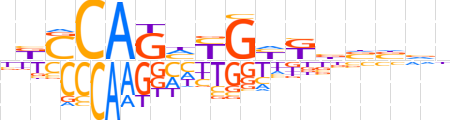 | ||||||||
| LOGO (reverse complement) | 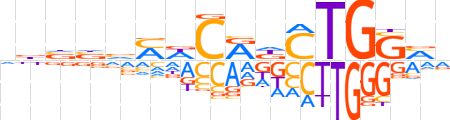 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 16 | ||||||||
| Quality | C | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | bKCCAGMYGWKbvbvn | ||||||||
| Best auROC (human) | 0.909 | ||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | 4 | ||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 321 | ||||||||
| TF family | More than 3 adjacent zinc finger factors {2.3.3} | ||||||||
| TF subfamily | ZNF238-like factors {2.3.3.16} | ||||||||
| HGNC | HGNC:13030 | ||||||||
| EntrezGene | GeneID:10472 (SSTAR profile) | ||||||||
| UniProt ID | ZBT18_HUMAN | ||||||||
| UniProt AC | Q99592 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | ZBTB18 expression | ||||||||
| ReMap ChIP-seq dataset list | ZBTB18 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 3.0 | 4.0 | 18.0 | 18.0 | 6.0 | 8.0 | 8.0 | 30.0 | 3.0 | 17.0 | 21.0 | 7.0 | 2.0 | 7.0 | 4.0 | 92.0 |
| 02 | 1.0 | 9.0 | 4.0 | 0.0 | 6.0 | 22.0 | 8.0 | 0.0 | 5.0 | 35.0 | 11.0 | 0.0 | 10.0 | 124.0 | 13.0 | 0.0 |
| 03 | 0.0 | 22.0 | 0.0 | 0.0 | 0.0 | 190.0 | 0.0 | 0.0 | 0.0 | 36.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 04 | 0.0 | 0.0 | 0.0 | 0.0 | 245.0 | 0.0 | 0.0 | 3.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 05 | 0.0 | 0.0 | 180.0 | 65.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.0 | 0.0 |
| 06 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 77.0 | 104.0 | 1.0 | 1.0 | 0.0 | 0.0 | 0.0 | 65.0 |
| 07 | 0.0 | 1.0 | 10.0 | 66.0 | 1.0 | 2.0 | 7.0 | 94.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 61.0 | 2.0 | 3.0 |
| 08 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 25.0 | 39.0 | 0.0 | 0.0 | 0.0 | 20.0 | 0.0 | 0.0 | 2.0 | 161.0 | 0.0 |
| 09 | 0.0 | 0.0 | 0.0 | 0.0 | 18.0 | 2.0 | 0.0 | 7.0 | 38.0 | 54.0 | 4.0 | 125.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 10 | 0.0 | 0.0 | 10.0 | 46.0 | 1.0 | 7.0 | 22.0 | 26.0 | 0.0 | 1.0 | 3.0 | 0.0 | 2.0 | 5.0 | 68.0 | 57.0 |
| 11 | 0.0 | 0.0 | 2.0 | 1.0 | 3.0 | 3.0 | 7.0 | 0.0 | 8.0 | 32.0 | 49.0 | 14.0 | 8.0 | 18.0 | 25.0 | 78.0 |
| 12 | 4.0 | 5.0 | 8.0 | 2.0 | 7.0 | 24.0 | 12.0 | 10.0 | 18.0 | 30.0 | 27.0 | 8.0 | 1.0 | 76.0 | 10.0 | 6.0 |
| 13 | 3.0 | 14.0 | 11.0 | 2.0 | 14.0 | 92.0 | 12.0 | 17.0 | 7.0 | 21.0 | 20.0 | 9.0 | 2.0 | 8.0 | 12.0 | 4.0 |
| 14 | 5.0 | 5.0 | 16.0 | 0.0 | 81.0 | 21.0 | 20.0 | 13.0 | 9.0 | 18.0 | 22.0 | 6.0 | 7.0 | 13.0 | 9.0 | 3.0 |
| 15 | 8.0 | 8.0 | 22.0 | 64.0 | 18.0 | 24.0 | 8.0 | 7.0 | 7.0 | 30.0 | 20.0 | 10.0 | 0.0 | 10.0 | 6.0 | 6.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -1.555 | -1.294 | 0.147 | 0.147 | -0.915 | -0.641 | -0.641 | 0.65 | -1.555 | 0.09 | 0.298 | -0.769 | -1.911 | -0.769 | -1.294 | 1.763 |
| 02 | -2.467 | -0.528 | -1.294 | -3.828 | -0.915 | 0.344 | -0.641 | -3.828 | -1.087 | 0.802 | -0.334 | -3.828 | -0.426 | 2.06 | -0.172 | -3.828 |
| 03 | -3.828 | 0.344 | -3.828 | -3.828 | -3.828 | 2.486 | -3.828 | -3.828 | -3.828 | 0.83 | -3.828 | -3.828 | -3.828 | -3.828 | -3.828 | -3.828 |
| 04 | -3.828 | -3.828 | -3.828 | -3.828 | 2.74 | -3.828 | -3.828 | -1.555 | -3.828 | -3.828 | -3.828 | -3.828 | -3.828 | -3.828 | -3.828 | -3.828 |
| 05 | -3.828 | -3.828 | 2.432 | 1.417 | -3.828 | -3.828 | -3.828 | -3.828 | -3.828 | -3.828 | -3.828 | -3.828 | -3.828 | -3.828 | -1.555 | -3.828 |
| 06 | -3.828 | -3.828 | -3.828 | -3.828 | -3.828 | -3.828 | -3.828 | -3.828 | 1.585 | 1.885 | -2.467 | -2.467 | -3.828 | -3.828 | -3.828 | 1.417 |
| 07 | -3.828 | -2.467 | -0.426 | 1.432 | -2.467 | -1.911 | -0.769 | 1.784 | -3.828 | -3.828 | -2.467 | -3.828 | -3.828 | 1.354 | -1.911 | -1.555 |
| 08 | -3.828 | -3.828 | -2.467 | -3.828 | -3.828 | 0.47 | 0.91 | -3.828 | -3.828 | -3.828 | 0.25 | -3.828 | -3.828 | -1.911 | 2.321 | -3.828 |
| 09 | -3.828 | -3.828 | -3.828 | -3.828 | 0.147 | -1.911 | -3.828 | -0.769 | 0.884 | 1.233 | -1.294 | 2.068 | -3.828 | -3.828 | -3.828 | -3.828 |
| 10 | -3.828 | -3.828 | -0.426 | 1.073 | -2.467 | -0.769 | 0.344 | 0.508 | -3.828 | -2.467 | -1.555 | -3.828 | -1.911 | -1.087 | 1.462 | 1.286 |
| 11 | -3.828 | -3.828 | -1.911 | -2.467 | -1.555 | -1.555 | -0.769 | -3.828 | -0.641 | 0.714 | 1.136 | -0.099 | -0.641 | 0.147 | 0.47 | 1.598 |
| 12 | -1.294 | -1.087 | -0.641 | -1.911 | -0.769 | 0.429 | -0.25 | -0.426 | 0.147 | 0.65 | 0.546 | -0.641 | -2.467 | 1.572 | -0.426 | -0.915 |
| 13 | -1.555 | -0.099 | -0.334 | -1.911 | -0.099 | 1.763 | -0.25 | 0.09 | -0.769 | 0.298 | 0.25 | -0.528 | -1.911 | -0.641 | -0.25 | -1.294 |
| 14 | -1.087 | -1.087 | 0.031 | -3.828 | 1.636 | 0.298 | 0.25 | -0.172 | -0.528 | 0.147 | 0.344 | -0.915 | -0.769 | -0.172 | -0.528 | -1.555 |
| 15 | -0.641 | -0.641 | 0.344 | 1.401 | 0.147 | 0.429 | -0.641 | -0.769 | -0.769 | 0.65 | 0.25 | -0.426 | -3.828 | -0.426 | -0.915 | -0.915 |