Model info
| Transcription factor | ZFP82 (GeneCards) | ||||||||
| Model | ZFP82_HUMAN.H11MO.0.C | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 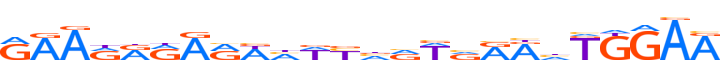 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 24 | ||||||||
| Quality | C | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | YTCChdYYbWbbRWdWYTYYYTYY | ||||||||
| Best auROC (human) | 0.93 | ||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | 3 | ||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 500 | ||||||||
| TF family | More than 3 adjacent zinc finger factors {2.3.3} | ||||||||
| TF subfamily | ZFP30-like factors {2.3.3.63} | ||||||||
| HGNC | HGNC:28682 | ||||||||
| EntrezGene | GeneID:284406 (SSTAR profile) | ||||||||
| UniProt ID | ZFP82_HUMAN | ||||||||
| UniProt AC | Q8N141 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | ZFP82 expression | ||||||||
| ReMap ChIP-seq dataset list | ZFP82 datasets | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 16.0 | 57.0 | 29.0 | 396.0 |
| 02 | 7.0 | 24.0 | 12.0 | 455.0 |
| 03 | 5.0 | 406.0 | 7.0 | 80.0 |
| 04 | 53.0 | 395.0 | 7.0 | 43.0 |
| 05 | 392.0 | 38.0 | 30.0 | 38.0 |
| 06 | 103.0 | 29.0 | 164.0 | 202.0 |
| 07 | 45.0 | 56.0 | 36.0 | 361.0 |
| 08 | 18.0 | 121.0 | 34.0 | 325.0 |
| 09 | 35.0 | 281.0 | 81.0 | 101.0 |
| 10 | 348.0 | 51.0 | 32.0 | 67.0 |
| 11 | 33.0 | 249.0 | 43.0 | 173.0 |
| 12 | 61.0 | 62.0 | 114.0 | 261.0 |
| 13 | 305.0 | 72.0 | 92.0 | 29.0 |
| 14 | 311.0 | 63.0 | 24.0 | 100.0 |
| 15 | 71.0 | 39.0 | 105.0 | 283.0 |
| 16 | 86.0 | 58.0 | 31.0 | 323.0 |
| 17 | 14.0 | 260.0 | 34.0 | 190.0 |
| 18 | 16.0 | 107.0 | 4.0 | 371.0 |
| 19 | 54.0 | 313.0 | 13.0 | 118.0 |
| 20 | 33.0 | 139.0 | 28.0 | 298.0 |
| 21 | 94.0 | 292.0 | 13.0 | 99.0 |
| 22 | 11.0 | 58.0 | 12.0 | 417.0 |
| 23 | 16.0 | 95.0 | 12.0 | 375.0 |
| 24 | 11.0 | 333.0 | 19.0 | 135.0 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -1.971 | -0.767 | -1.417 | 1.149 |
| 02 | -2.69 | -1.596 | -2.23 | 1.287 |
| 03 | -2.957 | 1.173 | -2.69 | -0.435 |
| 04 | -0.838 | 1.146 | -2.69 | -1.04 |
| 05 | 1.139 | -1.159 | -1.385 | -1.159 |
| 06 | -0.187 | -1.417 | 0.273 | 0.479 |
| 07 | -0.996 | -0.784 | -1.211 | 1.056 |
| 08 | -1.864 | -0.028 | -1.266 | 0.952 |
| 09 | -1.238 | 0.807 | -0.423 | -0.206 |
| 10 | 1.02 | -0.875 | -1.324 | -0.609 |
| 11 | -1.294 | 0.687 | -1.04 | 0.326 |
| 12 | -0.701 | -0.685 | -0.087 | 0.734 |
| 13 | 0.889 | -0.539 | -0.298 | -1.417 |
| 14 | 0.908 | -0.669 | -1.596 | -0.216 |
| 15 | -0.552 | -1.134 | -0.168 | 0.814 |
| 16 | -0.364 | -0.75 | -1.354 | 0.946 |
| 17 | -2.092 | 0.73 | -1.266 | 0.418 |
| 18 | -1.971 | -0.149 | -3.122 | 1.084 |
| 19 | -0.819 | 0.914 | -2.159 | -0.053 |
| 20 | -1.294 | 0.109 | -1.451 | 0.866 |
| 21 | -0.277 | 0.845 | -2.159 | -0.226 |
| 22 | -2.307 | -0.75 | -2.23 | 1.2 |
| 23 | -1.971 | -0.267 | -2.23 | 1.094 |
| 24 | -2.307 | 0.976 | -1.814 | 0.08 |