Model info
| Transcription factor | ZFX (GeneCards) | ||||||||
| Model | ZFX_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 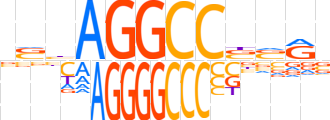 | ||||||||
| LOGO (reverse complement) | 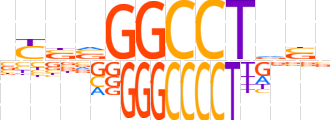 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 12 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nShAGGCCbvRv | ||||||||
| Best auROC (human) | 0.84 | ||||||||
| Best auROC (mouse) | 0.993 | ||||||||
| Peak sets in benchmark (human) | 10 | ||||||||
| Peak sets in benchmark (mouse) | 4 | ||||||||
| Aligned words | 500 | ||||||||
| TF family | More than 3 adjacent zinc finger factors {2.3.3} | ||||||||
| TF subfamily | ZFX/ZFY factors {2.3.3.65} | ||||||||
| HGNC | HGNC:12869 | ||||||||
| EntrezGene | GeneID:7543 (SSTAR profile) | ||||||||
| UniProt ID | ZFX_HUMAN | ||||||||
| UniProt AC | P17010 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | ZFX expression | ||||||||
| ReMap ChIP-seq dataset list | ZFX datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 7.0 | 40.0 | 42.0 | 1.0 | 24.0 | 87.0 | 60.0 | 6.0 | 13.0 | 95.0 | 43.0 | 1.0 | 7.0 | 15.0 | 46.0 | 11.0 |
| 02 | 12.0 | 26.0 | 10.0 | 3.0 | 37.0 | 107.0 | 31.0 | 62.0 | 37.0 | 77.0 | 26.0 | 51.0 | 3.0 | 6.0 | 1.0 | 9.0 |
| 03 | 87.0 | 0.0 | 0.0 | 2.0 | 216.0 | 0.0 | 0.0 | 0.0 | 68.0 | 0.0 | 0.0 | 0.0 | 125.0 | 0.0 | 0.0 | 0.0 |
| 04 | 0.0 | 0.0 | 496.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 |
| 05 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 496.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 06 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.0 | 493.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 |
| 07 | 0.0 | 3.0 | 0.0 | 0.0 | 4.0 | 486.0 | 3.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 08 | 0.0 | 1.0 | 3.0 | 0.0 | 6.0 | 211.0 | 159.0 | 113.0 | 0.0 | 0.0 | 3.0 | 0.0 | 0.0 | 1.0 | 1.0 | 0.0 |
| 09 | 3.0 | 1.0 | 2.0 | 0.0 | 59.0 | 67.0 | 71.0 | 16.0 | 12.0 | 117.0 | 34.0 | 3.0 | 5.0 | 68.0 | 37.0 | 3.0 |
| 10 | 16.0 | 3.0 | 59.0 | 1.0 | 95.0 | 2.0 | 144.0 | 12.0 | 55.0 | 6.0 | 80.0 | 3.0 | 6.0 | 3.0 | 12.0 | 1.0 |
| 11 | 27.0 | 46.0 | 91.0 | 8.0 | 1.0 | 3.0 | 10.0 | 0.0 | 46.0 | 111.0 | 121.0 | 17.0 | 1.0 | 8.0 | 4.0 | 4.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -1.451 | 0.248 | 0.296 | -3.122 | -0.256 | 1.02 | 0.65 | -1.596 | -0.856 | 1.108 | 0.32 | -3.122 | -1.451 | -0.717 | 0.387 | -1.018 |
| 02 | -0.934 | -0.177 | -1.11 | -2.23 | 0.171 | 1.226 | -0.004 | 0.683 | 0.171 | 0.898 | -0.177 | 0.489 | -2.23 | -1.596 | -3.122 | -1.211 |
| 03 | 1.02 | -4.397 | -4.397 | -2.58 | 1.927 | -4.397 | -4.397 | -4.397 | 0.775 | -4.397 | -4.397 | -4.397 | 1.381 | -4.397 | -4.397 | -4.397 |
| 04 | -4.397 | -4.397 | 2.757 | -4.397 | -4.397 | -4.397 | -4.397 | -4.397 | -4.397 | -4.397 | -4.397 | -4.397 | -4.397 | -4.397 | -2.58 | -4.397 |
| 05 | -4.397 | -4.397 | -4.397 | -4.397 | -4.397 | -4.397 | -4.397 | -4.397 | -3.122 | -4.397 | 2.757 | -3.122 | -4.397 | -4.397 | -4.397 | -4.397 |
| 06 | -4.397 | -3.122 | -4.397 | -4.397 | -4.397 | -4.397 | -4.397 | -4.397 | -2.23 | 2.751 | -4.397 | -4.397 | -4.397 | -3.122 | -4.397 | -4.397 |
| 07 | -4.397 | -2.23 | -4.397 | -4.397 | -1.971 | 2.737 | -2.23 | -2.58 | -4.397 | -4.397 | -4.397 | -4.397 | -4.397 | -4.397 | -4.397 | -4.397 |
| 08 | -4.397 | -3.122 | -2.23 | -4.397 | -1.596 | 1.903 | 1.621 | 1.28 | -4.397 | -4.397 | -2.23 | -4.397 | -4.397 | -3.122 | -3.122 | -4.397 |
| 09 | -2.23 | -3.122 | -2.58 | -4.397 | 0.634 | 0.76 | 0.818 | -0.654 | -0.934 | 1.315 | 0.087 | -2.23 | -1.766 | 0.775 | 0.171 | -2.23 |
| 10 | -0.654 | -2.23 | 0.634 | -3.122 | 1.108 | -2.58 | 1.522 | -0.934 | 0.564 | -1.596 | 0.936 | -2.23 | -1.596 | -2.23 | -0.934 | -3.122 |
| 11 | -0.14 | 0.387 | 1.065 | -1.324 | -3.122 | -2.23 | -1.11 | -4.397 | 0.387 | 1.263 | 1.349 | -0.595 | -3.122 | -1.324 | -1.971 | -1.971 |