Model info
| Transcription factor | ZIC1 (GeneCards) | ||||||||
| Model | ZIC1_HUMAN.H11MO.0.B | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO | 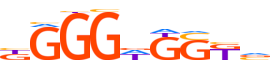 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | Integrative | ||||||||
| Model release | HOCOMOCOv9 | ||||||||
| Model length | 9 | ||||||||
| Quality | B | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | KGGGWGSKv | ||||||||
| Best auROC (human) | |||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | |||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 34 | ||||||||
| TF family | More than 3 adjacent zinc finger factors {2.3.3} | ||||||||
| TF subfamily | GLI-like factors {2.3.3.1} | ||||||||
| HGNC | HGNC:12872 | ||||||||
| EntrezGene | GeneID:7545 (SSTAR profile) | ||||||||
| UniProt ID | ZIC1_HUMAN | ||||||||
| UniProt AC | Q15915 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | ZIC1 expression | ||||||||
| ReMap ChIP-seq dataset list | ZIC1 datasets | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 1.571 | 4.387 | 17.276 | 10.019 |
| 02 | 2.924 | 1.95 | 28.378 | 0.0 |
| 03 | 0.0 | 0.975 | 31.302 | 0.975 |
| 04 | 0.0 | 1.95 | 31.302 | 0.0 |
| 05 | 9.965 | 0.975 | 5.849 | 16.463 |
| 06 | 3.899 | 0.0 | 25.453 | 3.899 |
| 07 | 1.95 | 7.798 | 23.504 | 0.0 |
| 08 | 1.083 | 2.924 | 8.882 | 20.363 |
| 09 | 7.176 | 14.758 | 9.125 | 2.193 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -1.323 | -0.557 | 0.681 | 0.17 |
| 02 | -0.883 | -1.179 | 1.158 | -2.35 |
| 03 | -2.35 | -1.602 | 1.253 | -1.602 |
| 04 | -2.35 | -1.179 | 1.253 | -2.35 |
| 05 | 0.165 | -1.602 | -0.312 | 0.635 |
| 06 | -0.655 | -2.35 | 1.053 | -0.655 |
| 07 | -1.179 | -0.058 | 0.976 | -2.35 |
| 08 | -1.545 | -0.883 | 0.06 | 0.838 |
| 09 | -0.132 | 0.531 | 0.085 | -1.097 |