Model info
| Transcription factor | ZIM3 (GeneCards) | ||||||||
| Model | ZIM3_HUMAN.H11DI.0.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 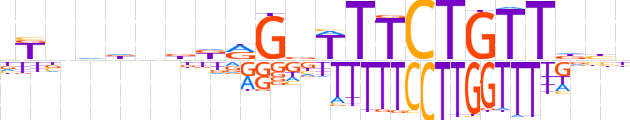 | ||||||||
| LOGO (reverse complement) | 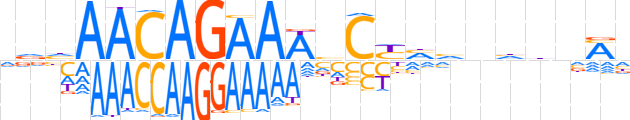 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 22 | ||||||||
| Quality | C | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | dKnddnbdRGbTTTCTGTTdhh | ||||||||
| Best auROC (human) | 0.989 | ||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | 3 | ||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 471 | ||||||||
| TF family | More than 3 adjacent zinc finger factors {2.3.3} | ||||||||
| TF subfamily | unclassified {2.3.3.0} | ||||||||
| HGNC | HGNC:16366 | ||||||||
| EntrezGene | GeneID:114026 (SSTAR profile) | ||||||||
| UniProt ID | ZIM3_HUMAN | ||||||||
| UniProt AC | Q96PE6 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | ZIM3 expression | ||||||||
| ReMap ChIP-seq dataset list | ZIM3 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 6.0 | 11.0 | 11.0 | 60.0 | 4.0 | 13.0 | 1.0 | 52.0 | 4.0 | 11.0 | 10.0 | 91.0 | 3.0 | 16.0 | 39.0 | 133.0 |
| 02 | 4.0 | 2.0 | 3.0 | 8.0 | 19.0 | 4.0 | 7.0 | 21.0 | 23.0 | 4.0 | 14.0 | 20.0 | 50.0 | 92.0 | 96.0 | 98.0 |
| 03 | 21.0 | 9.0 | 41.0 | 25.0 | 71.0 | 6.0 | 2.0 | 23.0 | 53.0 | 11.0 | 15.0 | 41.0 | 30.0 | 34.0 | 38.0 | 45.0 |
| 04 | 24.0 | 30.0 | 44.0 | 77.0 | 24.0 | 6.0 | 5.0 | 25.0 | 15.0 | 18.0 | 11.0 | 52.0 | 15.0 | 17.0 | 27.0 | 75.0 |
| 05 | 9.0 | 30.0 | 12.0 | 27.0 | 35.0 | 11.0 | 2.0 | 23.0 | 37.0 | 18.0 | 15.0 | 17.0 | 71.0 | 39.0 | 35.0 | 84.0 |
| 06 | 11.0 | 19.0 | 58.0 | 64.0 | 19.0 | 14.0 | 4.0 | 61.0 | 10.0 | 9.0 | 11.0 | 34.0 | 15.0 | 27.0 | 51.0 | 58.0 |
| 07 | 7.0 | 8.0 | 25.0 | 15.0 | 17.0 | 7.0 | 3.0 | 42.0 | 16.0 | 12.0 | 22.0 | 74.0 | 15.0 | 11.0 | 65.0 | 126.0 |
| 08 | 12.0 | 1.0 | 39.0 | 3.0 | 24.0 | 1.0 | 3.0 | 10.0 | 42.0 | 2.0 | 64.0 | 7.0 | 120.0 | 11.0 | 112.0 | 14.0 |
| 09 | 4.0 | 1.0 | 186.0 | 7.0 | 1.0 | 0.0 | 6.0 | 8.0 | 4.0 | 0.0 | 203.0 | 11.0 | 0.0 | 0.0 | 34.0 | 0.0 |
| 10 | 3.0 | 1.0 | 5.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 80.0 | 79.0 | 182.0 | 88.0 | 2.0 | 9.0 | 15.0 | 0.0 |
| 11 | 21.0 | 10.0 | 7.0 | 48.0 | 12.0 | 2.0 | 0.0 | 75.0 | 26.0 | 9.0 | 2.0 | 165.0 | 1.0 | 2.0 | 3.0 | 82.0 |
| 12 | 0.0 | 0.0 | 0.0 | 60.0 | 1.0 | 0.0 | 0.0 | 22.0 | 0.0 | 0.0 | 0.0 | 12.0 | 0.0 | 2.0 | 0.0 | 368.0 |
| 13 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.0 | 6.0 | 29.0 | 423.0 |
| 14 | 0.0 | 4.0 | 0.0 | 0.0 | 0.0 | 7.0 | 0.0 | 0.0 | 0.0 | 29.0 | 0.0 | 0.0 | 0.0 | 425.0 | 0.0 | 0.0 |
| 15 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 465.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 16 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 9.0 | 440.0 | 15.0 |
| 17 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 9.0 | 16.0 | 3.0 | 0.0 | 421.0 | 0.0 | 0.0 | 0.0 | 15.0 |
| 18 | 0.0 | 0.0 | 0.0 | 16.0 | 0.0 | 0.0 | 0.0 | 3.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.0 | 0.0 | 443.0 |
| 19 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 100.0 | 38.0 | 201.0 | 123.0 |
| 20 | 5.0 | 77.0 | 8.0 | 11.0 | 11.0 | 17.0 | 0.0 | 10.0 | 26.0 | 93.0 | 23.0 | 59.0 | 18.0 | 38.0 | 27.0 | 42.0 |
| 21 | 7.0 | 18.0 | 6.0 | 29.0 | 22.0 | 69.0 | 3.0 | 131.0 | 14.0 | 16.0 | 13.0 | 15.0 | 11.0 | 30.0 | 13.0 | 68.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -1.529 | -0.95 | -0.95 | 0.718 | -1.905 | -0.789 | -3.058 | 0.576 | -1.905 | -0.95 | -1.042 | 1.132 | -2.164 | -0.586 | 0.291 | 1.511 |
| 02 | -1.905 | -2.514 | -2.164 | -1.256 | -0.418 | -1.905 | -1.383 | -0.32 | -0.231 | -1.905 | -0.716 | -0.368 | 0.537 | 1.143 | 1.186 | 1.206 |
| 03 | -0.32 | -1.144 | 0.34 | -0.148 | 0.886 | -1.529 | -2.514 | -0.231 | 0.595 | -0.95 | -0.649 | 0.34 | 0.031 | 0.155 | 0.265 | 0.433 |
| 04 | -0.189 | 0.031 | 0.41 | 0.966 | -0.189 | -1.529 | -1.699 | -0.148 | -0.649 | -0.471 | -0.95 | 0.576 | -0.649 | -0.527 | -0.073 | 0.94 |
| 05 | -1.144 | 0.031 | -0.866 | -0.073 | 0.184 | -0.95 | -2.514 | -0.231 | 0.239 | -0.471 | -0.649 | -0.527 | 0.886 | 0.291 | 0.184 | 1.053 |
| 06 | -0.95 | -0.418 | 0.684 | 0.782 | -0.418 | -0.716 | -1.905 | 0.735 | -1.042 | -1.144 | -0.95 | 0.155 | -0.649 | -0.073 | 0.557 | 0.684 |
| 07 | -1.383 | -1.256 | -0.148 | -0.649 | -0.527 | -1.383 | -2.164 | 0.364 | -0.586 | -0.866 | -0.274 | 0.927 | -0.649 | -0.95 | 0.798 | 1.457 |
| 08 | -0.866 | -3.058 | 0.291 | -2.164 | -0.189 | -3.058 | -2.164 | -1.042 | 0.364 | -2.514 | 0.782 | -1.383 | 1.408 | -0.95 | 1.339 | -0.716 |
| 09 | -1.905 | -3.058 | 1.845 | -1.383 | -3.058 | -4.34 | -1.529 | -1.256 | -1.905 | -4.34 | 1.933 | -0.95 | -4.34 | -4.34 | 0.155 | -4.34 |
| 10 | -2.164 | -3.058 | -1.699 | -4.34 | -3.058 | -4.34 | -4.34 | -4.34 | 1.004 | 0.992 | 1.824 | 1.099 | -2.514 | -1.144 | -0.649 | -4.34 |
| 11 | -0.32 | -1.042 | -1.383 | 0.497 | -0.866 | -2.514 | -4.34 | 0.94 | -0.11 | -1.144 | -2.514 | 1.726 | -3.058 | -2.514 | -2.164 | 1.029 |
| 12 | -4.34 | -4.34 | -4.34 | 0.718 | -3.058 | -4.34 | -4.34 | -0.274 | -4.34 | -4.34 | -4.34 | -0.866 | -4.34 | -2.514 | -4.34 | 2.527 |
| 13 | -4.34 | -4.34 | -4.34 | -3.058 | -4.34 | -3.058 | -4.34 | -3.058 | -4.34 | -4.34 | -4.34 | -4.34 | -1.905 | -1.529 | -0.002 | 2.666 |
| 14 | -4.34 | -1.905 | -4.34 | -4.34 | -4.34 | -1.383 | -4.34 | -4.34 | -4.34 | -0.002 | -4.34 | -4.34 | -4.34 | 2.67 | -4.34 | -4.34 |
| 15 | -4.34 | -4.34 | -4.34 | -4.34 | -4.34 | -4.34 | -4.34 | 2.76 | -4.34 | -4.34 | -4.34 | -4.34 | -4.34 | -4.34 | -4.34 | -4.34 |
| 16 | -4.34 | -4.34 | -4.34 | -4.34 | -4.34 | -4.34 | -4.34 | -4.34 | -4.34 | -4.34 | -4.34 | -4.34 | -3.058 | -1.144 | 2.705 | -0.649 |
| 17 | -4.34 | -4.34 | -4.34 | -3.058 | -4.34 | -4.34 | -4.34 | -1.144 | -0.586 | -2.164 | -4.34 | 2.661 | -4.34 | -4.34 | -4.34 | -0.649 |
| 18 | -4.34 | -4.34 | -4.34 | -0.586 | -4.34 | -4.34 | -4.34 | -2.164 | -4.34 | -4.34 | -4.34 | -4.34 | -4.34 | -2.164 | -4.34 | 2.712 |
| 19 | -4.34 | -4.34 | -4.34 | -4.34 | -3.058 | -4.34 | -4.34 | -2.514 | -4.34 | -4.34 | -4.34 | -4.34 | 1.226 | 0.265 | 1.923 | 1.433 |
| 20 | -1.699 | 0.966 | -1.256 | -0.95 | -0.95 | -0.527 | -4.34 | -1.042 | -0.11 | 1.154 | -0.231 | 0.701 | -0.471 | 0.265 | -0.073 | 0.364 |
| 21 | -1.383 | -0.471 | -1.529 | -0.002 | -0.274 | 0.857 | -2.164 | 1.496 | -0.716 | -0.586 | -0.789 | -0.649 | -0.95 | 0.031 | -0.789 | 0.843 |