Model info
| Transcription factor | ZNF263 (GeneCards) | ||||||||
| Model | ZN263_HUMAN.H11DI.0.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 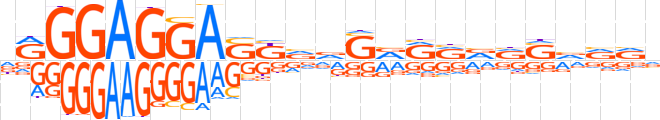 | ||||||||
| LOGO (reverse complement) | 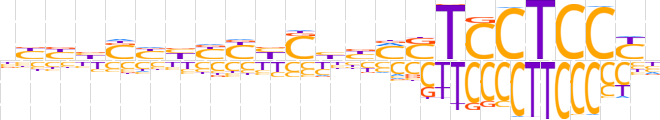 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 23 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | dRGGAGGASKvvSRRRRRSvvRv | ||||||||
| Best auROC (human) | 0.986 | ||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | 9 | ||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 467 | ||||||||
| TF family | More than 3 adjacent zinc finger factors {2.3.3} | ||||||||
| TF subfamily | unclassified {2.3.3.0} | ||||||||
| HGNC | HGNC:13056 | ||||||||
| EntrezGene | GeneID:10127 (SSTAR profile) | ||||||||
| UniProt ID | ZN263_HUMAN | ||||||||
| UniProt AC | O14978 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | ZNF263 expression | ||||||||
| ReMap ChIP-seq dataset list | ZNF263 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 25.0 | 4.0 | 108.0 | 3.0 | 17.0 | 0.0 | 28.0 | 5.0 | 82.0 | 3.0 | 87.0 | 6.0 | 2.0 | 1.0 | 47.0 | 5.0 |
| 02 | 0.0 | 0.0 | 126.0 | 0.0 | 0.0 | 0.0 | 8.0 | 0.0 | 3.0 | 2.0 | 257.0 | 8.0 | 0.0 | 0.0 | 19.0 | 0.0 |
| 03 | 0.0 | 0.0 | 3.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 4.0 | 1.0 | 403.0 | 2.0 | 0.0 | 0.0 | 8.0 | 0.0 |
| 04 | 4.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 416.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 |
| 05 | 5.0 | 0.0 | 408.0 | 9.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 06 | 0.0 | 0.0 | 5.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 58.0 | 350.0 | 1.0 | 0.0 | 0.0 | 8.0 | 1.0 |
| 07 | 0.0 | 0.0 | 0.0 | 0.0 | 57.0 | 1.0 | 0.0 | 0.0 | 356.0 | 7.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 0.0 |
| 08 | 22.0 | 112.0 | 273.0 | 7.0 | 0.0 | 0.0 | 6.0 | 2.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 09 | 1.0 | 0.0 | 20.0 | 1.0 | 15.0 | 17.0 | 35.0 | 45.0 | 7.0 | 23.0 | 223.0 | 27.0 | 0.0 | 1.0 | 7.0 | 1.0 |
| 10 | 3.0 | 5.0 | 13.0 | 2.0 | 9.0 | 13.0 | 15.0 | 4.0 | 106.0 | 19.0 | 152.0 | 8.0 | 13.0 | 34.0 | 22.0 | 5.0 |
| 11 | 41.0 | 7.0 | 81.0 | 2.0 | 22.0 | 21.0 | 14.0 | 14.0 | 114.0 | 19.0 | 48.0 | 21.0 | 4.0 | 2.0 | 12.0 | 1.0 |
| 12 | 12.0 | 14.0 | 147.0 | 8.0 | 8.0 | 11.0 | 24.0 | 6.0 | 7.0 | 9.0 | 137.0 | 2.0 | 0.0 | 5.0 | 27.0 | 6.0 |
| 13 | 7.0 | 5.0 | 12.0 | 3.0 | 19.0 | 7.0 | 7.0 | 6.0 | 201.0 | 19.0 | 96.0 | 19.0 | 3.0 | 5.0 | 13.0 | 1.0 |
| 14 | 13.0 | 19.0 | 188.0 | 10.0 | 12.0 | 5.0 | 15.0 | 4.0 | 36.0 | 11.0 | 71.0 | 10.0 | 1.0 | 3.0 | 20.0 | 5.0 |
| 15 | 5.0 | 1.0 | 52.0 | 4.0 | 7.0 | 8.0 | 15.0 | 8.0 | 45.0 | 34.0 | 196.0 | 19.0 | 1.0 | 9.0 | 13.0 | 6.0 |
| 16 | 8.0 | 3.0 | 40.0 | 7.0 | 25.0 | 8.0 | 16.0 | 3.0 | 173.0 | 24.0 | 68.0 | 11.0 | 8.0 | 8.0 | 19.0 | 2.0 |
| 17 | 18.0 | 18.0 | 161.0 | 17.0 | 13.0 | 4.0 | 18.0 | 8.0 | 41.0 | 22.0 | 67.0 | 13.0 | 4.0 | 2.0 | 10.0 | 7.0 |
| 18 | 7.0 | 8.0 | 58.0 | 3.0 | 7.0 | 6.0 | 21.0 | 12.0 | 26.0 | 29.0 | 186.0 | 15.0 | 2.0 | 6.0 | 32.0 | 5.0 |
| 19 | 8.0 | 10.0 | 23.0 | 1.0 | 14.0 | 13.0 | 11.0 | 11.0 | 182.0 | 26.0 | 70.0 | 19.0 | 5.0 | 11.0 | 11.0 | 8.0 |
| 20 | 27.0 | 26.0 | 147.0 | 9.0 | 16.0 | 12.0 | 17.0 | 15.0 | 50.0 | 13.0 | 51.0 | 1.0 | 5.0 | 5.0 | 23.0 | 6.0 |
| 21 | 19.0 | 12.0 | 63.0 | 4.0 | 23.0 | 6.0 | 18.0 | 9.0 | 51.0 | 19.0 | 152.0 | 16.0 | 5.0 | 8.0 | 15.0 | 3.0 |
| 22 | 14.0 | 11.0 | 58.0 | 15.0 | 19.0 | 11.0 | 6.0 | 9.0 | 102.0 | 43.0 | 78.0 | 25.0 | 4.0 | 7.0 | 17.0 | 4.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.055 | -1.812 | 1.397 | -2.072 | -0.434 | -4.262 | 0.057 | -1.607 | 1.122 | -2.072 | 1.181 | -1.436 | -2.423 | -2.968 | 0.569 | -1.607 |
| 02 | -4.262 | -4.262 | 1.55 | -4.262 | -4.262 | -4.262 | -1.163 | -4.262 | -2.072 | -2.423 | 2.262 | -1.163 | -4.262 | -4.262 | -0.325 | -4.262 |
| 03 | -4.262 | -4.262 | -2.072 | -4.262 | -4.262 | -4.262 | -2.423 | -4.262 | -1.812 | -2.968 | 2.711 | -2.423 | -4.262 | -4.262 | -1.163 | -4.262 |
| 04 | -1.812 | -4.262 | -4.262 | -4.262 | -2.968 | -4.262 | -4.262 | -4.262 | 2.743 | -4.262 | -4.262 | -4.262 | -2.968 | -4.262 | -2.968 | -4.262 |
| 05 | -1.607 | -4.262 | 2.723 | -1.051 | -4.262 | -4.262 | -4.262 | -4.262 | -4.262 | -4.262 | -2.968 | -4.262 | -4.262 | -4.262 | -4.262 | -4.262 |
| 06 | -4.262 | -4.262 | -1.607 | -4.262 | -4.262 | -4.262 | -4.262 | -4.262 | -4.262 | 0.778 | 2.57 | -2.968 | -4.262 | -4.262 | -1.163 | -2.968 |
| 07 | -4.262 | -4.262 | -4.262 | -4.262 | 0.761 | -2.968 | -4.262 | -4.262 | 2.587 | -1.29 | -4.262 | -4.262 | -2.968 | -4.262 | -2.968 | -4.262 |
| 08 | -0.181 | 1.433 | 2.322 | -1.29 | -4.262 | -4.262 | -1.436 | -2.423 | -4.262 | -4.262 | -2.968 | -4.262 | -4.262 | -4.262 | -4.262 | -4.262 |
| 09 | -2.968 | -4.262 | -0.275 | -2.968 | -0.556 | -0.434 | 0.277 | 0.526 | -1.29 | -0.137 | 2.12 | 0.021 | -4.262 | -2.968 | -1.29 | -2.968 |
| 10 | -2.072 | -1.607 | -0.695 | -2.423 | -1.051 | -0.695 | -0.556 | -1.812 | 1.378 | -0.325 | 1.737 | -1.163 | -0.695 | 0.248 | -0.181 | -1.607 |
| 11 | 0.434 | -1.29 | 1.11 | -2.423 | -0.181 | -0.227 | -0.623 | -0.623 | 1.451 | -0.325 | 0.59 | -0.227 | -1.812 | -2.423 | -0.773 | -2.968 |
| 12 | -0.773 | -0.623 | 1.704 | -1.163 | -1.163 | -0.857 | -0.095 | -1.436 | -1.29 | -1.051 | 1.634 | -2.423 | -4.262 | -1.607 | 0.021 | -1.436 |
| 13 | -1.29 | -1.607 | -0.773 | -2.072 | -0.325 | -1.29 | -1.29 | -1.436 | 2.016 | -0.325 | 1.279 | -0.325 | -2.072 | -1.607 | -0.695 | -2.968 |
| 14 | -0.695 | -0.325 | 1.949 | -0.949 | -0.773 | -1.607 | -0.556 | -1.812 | 0.305 | -0.857 | 0.979 | -0.949 | -2.968 | -2.072 | -0.275 | -1.607 |
| 15 | -1.607 | -2.968 | 0.67 | -1.812 | -1.29 | -1.163 | -0.556 | -1.163 | 0.526 | 0.248 | 1.991 | -0.325 | -2.968 | -1.051 | -0.695 | -1.436 |
| 16 | -1.163 | -2.072 | 0.409 | -1.29 | -0.055 | -1.163 | -0.493 | -2.072 | 1.866 | -0.095 | 0.936 | -0.857 | -1.163 | -1.163 | -0.325 | -2.423 |
| 17 | -0.378 | -0.378 | 1.795 | -0.434 | -0.695 | -1.812 | -0.378 | -1.163 | 0.434 | -0.181 | 0.921 | -0.695 | -1.812 | -2.423 | -0.949 | -1.29 |
| 18 | -1.29 | -1.163 | 0.778 | -2.072 | -1.29 | -1.436 | -0.227 | -0.773 | -0.016 | 0.091 | 1.939 | -0.556 | -2.423 | -1.436 | 0.188 | -1.607 |
| 19 | -1.163 | -0.949 | -0.137 | -2.968 | -0.623 | -0.695 | -0.857 | -0.857 | 1.917 | -0.016 | 0.965 | -0.325 | -1.607 | -0.857 | -0.857 | -1.163 |
| 20 | 0.021 | -0.016 | 1.704 | -1.051 | -0.493 | -0.773 | -0.434 | -0.556 | 0.631 | -0.695 | 0.65 | -2.968 | -1.607 | -1.607 | -0.137 | -1.436 |
| 21 | -0.325 | -0.773 | 0.86 | -1.812 | -0.137 | -1.436 | -0.378 | -1.051 | 0.65 | -0.325 | 1.737 | -0.493 | -1.607 | -1.163 | -0.556 | -2.072 |
| 22 | -0.623 | -0.857 | 0.778 | -0.556 | -0.325 | -0.857 | -1.436 | -1.051 | 1.34 | 0.481 | 1.073 | -0.055 | -1.812 | -1.29 | -0.434 | -1.812 |