Model info
| Transcription factor | ZNF320 (GeneCards) | ||||||||
| Model | ZN320_HUMAN.H11DI.0.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 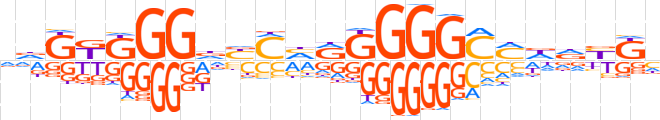 | ||||||||
| LOGO (reverse complement) | 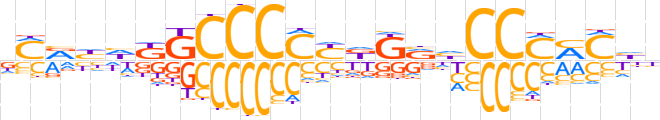 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 23 | ||||||||
| Quality | C | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | vdKKKGGdYMhRGGGGCMddYKh | ||||||||
| Best auROC (human) | 0.985 | ||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | 3 | ||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 458 | ||||||||
| TF family | More than 3 adjacent zinc finger factors {2.3.3} | ||||||||
| TF subfamily | unclassified {2.3.3.0} | ||||||||
| HGNC | HGNC:13842 | ||||||||
| EntrezGene | GeneID:162967 (SSTAR profile) | ||||||||
| UniProt ID | ZN320_HUMAN | ||||||||
| UniProt AC | A2RRD8 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | ZNF320 expression | ||||||||
| ReMap ChIP-seq dataset list | ZNF320 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 131.0 | 12.0 | 54.0 | 32.0 | 19.0 | 12.0 | 9.0 | 13.0 | 46.0 | 7.0 | 35.0 | 25.0 | 8.0 | 1.0 | 19.0 | 12.0 |
| 02 | 7.0 | 6.0 | 177.0 | 14.0 | 5.0 | 3.0 | 14.0 | 10.0 | 7.0 | 5.0 | 94.0 | 11.0 | 6.0 | 1.0 | 61.0 | 14.0 |
| 03 | 5.0 | 0.0 | 16.0 | 4.0 | 1.0 | 3.0 | 4.0 | 7.0 | 12.0 | 6.0 | 125.0 | 203.0 | 2.0 | 1.0 | 29.0 | 17.0 |
| 04 | 5.0 | 1.0 | 8.0 | 6.0 | 4.0 | 2.0 | 4.0 | 0.0 | 26.0 | 11.0 | 104.0 | 33.0 | 4.0 | 3.0 | 219.0 | 5.0 |
| 05 | 0.0 | 0.0 | 39.0 | 0.0 | 0.0 | 0.0 | 17.0 | 0.0 | 5.0 | 4.0 | 321.0 | 5.0 | 0.0 | 0.0 | 44.0 | 0.0 |
| 06 | 0.0 | 1.0 | 3.0 | 1.0 | 0.0 | 0.0 | 4.0 | 0.0 | 5.0 | 6.0 | 409.0 | 1.0 | 0.0 | 0.0 | 5.0 | 0.0 |
| 07 | 3.0 | 1.0 | 0.0 | 1.0 | 1.0 | 2.0 | 0.0 | 4.0 | 160.0 | 11.0 | 134.0 | 116.0 | 0.0 | 0.0 | 2.0 | 0.0 |
| 08 | 11.0 | 110.0 | 5.0 | 38.0 | 5.0 | 7.0 | 1.0 | 1.0 | 15.0 | 73.0 | 23.0 | 25.0 | 12.0 | 76.0 | 28.0 | 5.0 |
| 09 | 6.0 | 31.0 | 4.0 | 2.0 | 21.0 | 221.0 | 6.0 | 18.0 | 11.0 | 39.0 | 4.0 | 3.0 | 8.0 | 50.0 | 6.0 | 5.0 |
| 10 | 19.0 | 11.0 | 3.0 | 13.0 | 186.0 | 108.0 | 10.0 | 37.0 | 8.0 | 4.0 | 3.0 | 5.0 | 10.0 | 10.0 | 4.0 | 4.0 |
| 11 | 28.0 | 1.0 | 184.0 | 10.0 | 66.0 | 7.0 | 26.0 | 34.0 | 8.0 | 0.0 | 9.0 | 3.0 | 4.0 | 2.0 | 51.0 | 2.0 |
| 12 | 5.0 | 0.0 | 93.0 | 8.0 | 0.0 | 0.0 | 9.0 | 1.0 | 23.0 | 2.0 | 201.0 | 44.0 | 4.0 | 0.0 | 40.0 | 5.0 |
| 13 | 0.0 | 0.0 | 32.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 4.0 | 0.0 | 335.0 | 4.0 | 0.0 | 0.0 | 58.0 | 0.0 |
| 14 | 0.0 | 0.0 | 4.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 8.0 | 1.0 | 418.0 | 0.0 | 0.0 | 0.0 | 4.0 | 0.0 |
| 15 | 0.0 | 0.0 | 8.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 18.0 | 6.0 | 394.0 | 8.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 16 | 0.0 | 18.0 | 0.0 | 0.0 | 1.0 | 5.0 | 0.0 | 0.0 | 107.0 | 290.0 | 3.0 | 3.0 | 4.0 | 4.0 | 0.0 | 0.0 |
| 17 | 34.0 | 69.0 | 9.0 | 0.0 | 84.0 | 191.0 | 6.0 | 36.0 | 0.0 | 3.0 | 0.0 | 0.0 | 1.0 | 2.0 | 0.0 | 0.0 |
| 18 | 32.0 | 15.0 | 47.0 | 25.0 | 142.0 | 3.0 | 3.0 | 117.0 | 7.0 | 0.0 | 4.0 | 4.0 | 23.0 | 1.0 | 11.0 | 1.0 |
| 19 | 103.0 | 8.0 | 54.0 | 39.0 | 11.0 | 5.0 | 2.0 | 1.0 | 29.0 | 7.0 | 21.0 | 8.0 | 26.0 | 4.0 | 112.0 | 5.0 |
| 20 | 13.0 | 50.0 | 18.0 | 88.0 | 7.0 | 3.0 | 4.0 | 10.0 | 4.0 | 21.0 | 22.0 | 142.0 | 4.0 | 25.0 | 7.0 | 17.0 |
| 21 | 1.0 | 4.0 | 19.0 | 4.0 | 29.0 | 4.0 | 56.0 | 10.0 | 3.0 | 8.0 | 26.0 | 14.0 | 7.0 | 8.0 | 224.0 | 18.0 |
| 22 | 4.0 | 27.0 | 5.0 | 4.0 | 3.0 | 7.0 | 4.0 | 10.0 | 90.0 | 154.0 | 33.0 | 48.0 | 6.0 | 16.0 | 12.0 | 12.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 1.561 | -0.801 | 0.679 | 0.161 | -0.352 | -0.801 | -1.078 | -0.723 | 0.52 | -1.318 | 0.25 | -0.083 | -1.191 | -2.995 | -0.352 | -0.801 |
| 02 | -1.318 | -1.464 | 1.862 | -0.651 | -1.634 | -2.099 | -0.651 | -0.977 | -1.318 | -1.634 | 1.231 | -0.885 | -1.464 | -2.995 | 0.8 | -0.651 |
| 03 | -1.634 | -4.285 | -0.521 | -1.84 | -2.995 | -2.099 | -1.84 | -1.318 | -0.801 | -1.464 | 1.515 | 1.998 | -2.45 | -2.995 | 0.064 | -0.461 |
| 04 | -1.634 | -2.995 | -1.191 | -1.464 | -1.84 | -2.45 | -1.84 | -4.285 | -0.044 | -0.885 | 1.331 | 0.191 | -1.84 | -2.099 | 2.074 | -1.634 |
| 05 | -4.285 | -4.285 | 0.357 | -4.285 | -4.285 | -4.285 | -0.461 | -4.285 | -1.634 | -1.84 | 2.456 | -1.634 | -4.285 | -4.285 | 0.476 | -4.285 |
| 06 | -4.285 | -2.995 | -2.099 | -2.995 | -4.285 | -4.285 | -1.84 | -4.285 | -1.634 | -1.464 | 2.698 | -2.995 | -4.285 | -4.285 | -1.634 | -4.285 |
| 07 | -2.099 | -2.995 | -4.285 | -2.995 | -2.995 | -2.45 | -4.285 | -1.84 | 1.761 | -0.885 | 1.584 | 1.44 | -4.285 | -4.285 | -2.45 | -4.285 |
| 08 | -0.885 | 1.387 | -1.634 | 0.331 | -1.634 | -1.318 | -2.995 | -2.995 | -0.584 | 0.979 | -0.165 | -0.083 | -0.801 | 1.019 | 0.029 | -1.634 |
| 09 | -1.464 | 0.13 | -1.84 | -2.45 | -0.254 | 2.083 | -1.464 | -0.405 | -0.885 | 0.357 | -1.84 | -2.099 | -1.191 | 0.603 | -1.464 | -1.634 |
| 10 | -0.352 | -0.885 | -2.099 | -0.723 | 1.911 | 1.369 | -0.977 | 0.305 | -1.191 | -1.84 | -2.099 | -1.634 | -0.977 | -0.977 | -1.84 | -1.84 |
| 11 | 0.029 | -2.995 | 1.9 | -0.977 | 0.879 | -1.318 | -0.044 | 0.221 | -1.191 | -4.285 | -1.078 | -2.099 | -1.84 | -2.45 | 0.623 | -2.45 |
| 12 | -1.634 | -4.285 | 1.22 | -1.191 | -4.285 | -4.285 | -1.078 | -2.995 | -0.165 | -2.45 | 1.989 | 0.476 | -1.84 | -4.285 | 0.382 | -1.634 |
| 13 | -4.285 | -4.285 | 0.161 | -4.285 | -4.285 | -4.285 | -2.45 | -4.285 | -1.84 | -4.285 | 2.499 | -1.84 | -4.285 | -4.285 | 0.75 | -4.285 |
| 14 | -4.285 | -4.285 | -1.84 | -4.285 | -4.285 | -4.285 | -4.285 | -4.285 | -1.191 | -2.995 | 2.72 | -4.285 | -4.285 | -4.285 | -1.84 | -4.285 |
| 15 | -4.285 | -4.285 | -1.191 | -4.285 | -4.285 | -4.285 | -2.995 | -4.285 | -0.405 | -1.464 | 2.661 | -1.191 | -4.285 | -4.285 | -4.285 | -4.285 |
| 16 | -4.285 | -0.405 | -4.285 | -4.285 | -2.995 | -1.634 | -4.285 | -4.285 | 1.36 | 2.355 | -2.099 | -2.099 | -1.84 | -1.84 | -4.285 | -4.285 |
| 17 | 0.221 | 0.923 | -1.078 | -4.285 | 1.119 | 1.938 | -1.464 | 0.277 | -4.285 | -2.099 | -4.285 | -4.285 | -2.995 | -2.45 | -4.285 | -4.285 |
| 18 | 0.161 | -0.584 | 0.542 | -0.083 | 1.642 | -2.099 | -2.099 | 1.449 | -1.318 | -4.285 | -1.84 | -1.84 | -0.165 | -2.995 | -0.885 | -2.995 |
| 19 | 1.322 | -1.191 | 0.679 | 0.357 | -0.885 | -1.634 | -2.45 | -2.995 | 0.064 | -1.318 | -0.254 | -1.191 | -0.044 | -1.84 | 1.405 | -1.634 |
| 20 | -0.723 | 0.603 | -0.405 | 1.165 | -1.318 | -2.099 | -1.84 | -0.977 | -1.84 | -0.254 | -0.208 | 1.642 | -1.84 | -0.083 | -1.318 | -0.461 |
| 21 | -2.995 | -1.84 | -0.352 | -1.84 | 0.064 | -1.84 | 0.715 | -0.977 | -2.099 | -1.191 | -0.044 | -0.651 | -1.318 | -1.191 | 2.097 | -0.405 |
| 22 | -1.84 | -0.007 | -1.634 | -1.84 | -2.099 | -1.318 | -1.84 | -0.977 | 1.187 | 1.723 | 0.191 | 0.562 | -1.464 | -0.521 | -0.801 | -0.801 |