Model info
| Transcription factor | Znf335 | ||||||||
| Model | ZN335_MOUSE.H11MO.0.A | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO | 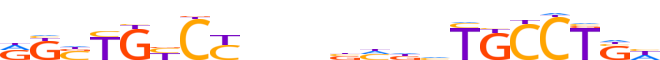 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 22 | ||||||||
| Quality | A | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | RKSTGYCYnnnbhdvTGCCTSh | ||||||||
| Best auROC (human) | 0.96 | ||||||||
| Best auROC (mouse) | 0.968 | ||||||||
| Peak sets in benchmark (human) | 2 | ||||||||
| Peak sets in benchmark (mouse) | 4 | ||||||||
| Aligned words | 103 | ||||||||
| TF family | Factors with multiple dispersed zinc fingers {2.3.4} | ||||||||
| TF subfamily | unclassified {2.3.4.0} | ||||||||
| MGI | MGI:2682313 | ||||||||
| EntrezGene | GeneID:329559 (SSTAR profile) | ||||||||
| UniProt ID | ZN335_MOUSE | ||||||||
| UniProt AC | A2A5K6 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | Znf335 expression | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 30.0 | 2.0 | 60.0 | 11.0 |
| 02 | 6.0 | 8.0 | 78.0 | 11.0 |
| 03 | 8.0 | 64.0 | 17.0 | 14.0 |
| 04 | 2.0 | 22.0 | 2.0 | 77.0 |
| 05 | 1.0 | 4.0 | 87.0 | 11.0 |
| 06 | 5.0 | 35.0 | 10.0 | 53.0 |
| 07 | 1.0 | 94.0 | 3.0 | 5.0 |
| 08 | 1.0 | 60.0 | 2.0 | 40.0 |
| 09 | 20.0 | 19.0 | 30.0 | 34.0 |
| 10 | 15.0 | 36.0 | 23.0 | 29.0 |
| 11 | 28.0 | 22.0 | 36.0 | 17.0 |
| 12 | 11.0 | 28.0 | 51.0 | 13.0 |
| 13 | 24.0 | 56.0 | 6.0 | 17.0 |
| 14 | 19.0 | 8.0 | 49.0 | 27.0 |
| 15 | 28.0 | 46.0 | 16.0 | 13.0 |
| 16 | 3.0 | 14.0 | 0.0 | 86.0 |
| 17 | 2.0 | 4.0 | 84.0 | 13.0 |
| 18 | 2.0 | 90.0 | 1.0 | 10.0 |
| 19 | 4.0 | 95.0 | 0.0 | 4.0 |
| 20 | 4.0 | 5.0 | 5.0 | 89.0 |
| 21 | 10.0 | 11.0 | 72.0 | 10.0 |
| 22 | 49.0 | 16.0 | 4.0 | 34.0 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.147 | -2.142 | 0.821 | -0.794 |
| 02 | -1.324 | -1.078 | 1.079 | -0.794 |
| 03 | -1.078 | 0.884 | -0.393 | -0.574 |
| 04 | -2.142 | -0.15 | -2.142 | 1.066 |
| 05 | -2.523 | -1.652 | 1.187 | -0.794 |
| 06 | -1.475 | 0.295 | -0.88 | 0.699 |
| 07 | -2.523 | 1.263 | -1.867 | -1.475 |
| 08 | -2.523 | 0.821 | -2.142 | 0.425 |
| 09 | -0.24 | -0.289 | 0.147 | 0.267 |
| 10 | -0.51 | 0.323 | -0.108 | 0.114 |
| 11 | 0.08 | -0.15 | 0.323 | -0.393 |
| 12 | -0.794 | 0.08 | 0.662 | -0.642 |
| 13 | -0.067 | 0.753 | -1.324 | -0.393 |
| 14 | -0.289 | -1.078 | 0.623 | 0.045 |
| 15 | 0.08 | 0.561 | -0.45 | -0.642 |
| 16 | -1.867 | -0.574 | -3.145 | 1.175 |
| 17 | -2.142 | -1.652 | 1.152 | -0.642 |
| 18 | -2.142 | 1.22 | -2.523 | -0.88 |
| 19 | -1.652 | 1.274 | -3.145 | -1.652 |
| 20 | -1.652 | -1.475 | -1.475 | 1.209 |
| 21 | -0.88 | -0.794 | 1.0 | -0.88 |
| 22 | 0.623 | -0.45 | -1.652 | 0.267 |