Model info
| Transcription factor | ZNF350 (GeneCards) | ||||||||
| Model | ZN350_HUMAN.H11MO.1.D | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 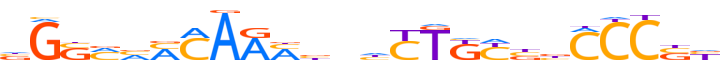 | ||||||||
| Data source | Integrative | ||||||||
| Model release | HOCOMOCOv9 | ||||||||
| Model length | 24 | ||||||||
| Quality | D | ||||||||
| Motif rank | 1 | ||||||||
| Consensus | nSGGGvSRMAGnnvKTTGKbKSCn | ||||||||
| Best auROC (human) | 0.671 | ||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | 3 | ||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 15 | ||||||||
| TF family | More than 3 adjacent zinc finger factors {2.3.3} | ||||||||
| TF subfamily | ZNF350-like factors {2.3.3.30} | ||||||||
| HGNC | HGNC:16656 | ||||||||
| EntrezGene | GeneID:59348 (SSTAR profile) | ||||||||
| UniProt ID | ZN350_HUMAN | ||||||||
| UniProt AC | Q9GZX5 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | ZNF350 expression | ||||||||
| ReMap ChIP-seq dataset list | ZNF350 datasets | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 8.271 | 1.676 | 3.378 | 1.676 |
| 02 | 2.872 | 7.34 | 4.787 | 0.0 |
| 03 | 1.702 | 0.0 | 13.298 | 0.0 |
| 04 | 0.957 | 0.0 | 13.298 | 0.745 |
| 05 | 0.957 | 0.0 | 12.128 | 1.915 |
| 06 | 4.681 | 3.83 | 5.532 | 0.957 |
| 07 | 3.83 | 5.638 | 5.532 | 0.0 |
| 08 | 2.872 | 0.0 | 9.468 | 2.66 |
| 09 | 2.872 | 10.426 | 0.957 | 0.745 |
| 10 | 12.34 | 1.702 | 0.0 | 0.957 |
| 11 | 5.532 | 0.0 | 9.468 | 0.0 |
| 12 | 3.83 | 0.957 | 6.383 | 3.83 |
| 13 | 2.66 | 2.872 | 4.787 | 4.681 |
| 14 | 5.745 | 3.617 | 4.681 | 0.957 |
| 15 | 0.957 | 0.957 | 3.83 | 9.255 |
| 16 | 0.0 | 4.787 | 0.0 | 10.213 |
| 17 | 0.0 | 0.957 | 0.0 | 14.043 |
| 18 | 0.0 | 0.0 | 10.319 | 4.681 |
| 19 | 0.0 | 1.702 | 4.681 | 8.617 |
| 20 | 0.745 | 1.915 | 5.745 | 6.596 |
| 21 | 0.745 | 1.915 | 8.511 | 3.83 |
| 22 | 0.957 | 9.468 | 3.83 | 0.745 |
| 23 | 0.0 | 13.298 | 0.745 | 0.957 |
| 24 | 1.862 | 7.606 | 3.67 | 1.862 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.704 | -0.632 | -0.088 | -0.632 |
| 02 | -0.221 | 0.594 | 0.211 | -1.878 |
| 03 | -0.621 | -1.878 | 1.15 | -1.878 |
| 04 | -0.996 | -1.878 | 1.15 | -1.136 |
| 05 | -0.996 | -1.878 | 1.062 | -0.535 |
| 06 | 0.191 | 0.018 | 0.338 | -0.996 |
| 07 | 0.018 | 0.355 | 0.338 | -1.878 |
| 08 | -0.221 | -1.878 | 0.829 | -0.283 |
| 09 | -0.221 | 0.919 | -0.996 | -1.136 |
| 10 | 1.079 | -0.621 | -1.878 | -0.996 |
| 11 | 0.338 | -1.878 | 0.829 | -1.878 |
| 12 | 0.018 | -0.996 | 0.467 | 0.018 |
| 13 | -0.283 | -0.221 | 0.211 | 0.191 |
| 14 | 0.372 | -0.03 | 0.191 | -0.996 |
| 15 | -0.996 | -0.996 | 0.018 | 0.808 |
| 16 | -1.878 | 0.211 | -1.878 | 0.9 |
| 17 | -1.878 | -0.996 | -1.878 | 1.201 |
| 18 | -1.878 | -1.878 | 0.91 | 0.191 |
| 19 | -1.878 | -0.621 | 0.191 | 0.742 |
| 20 | -1.136 | -0.535 | 0.372 | 0.496 |
| 21 | -1.136 | -0.535 | 0.73 | 0.018 |
| 22 | -0.996 | 0.829 | 0.018 | -1.136 |
| 23 | -1.878 | 1.15 | -1.136 | -0.996 |
| 24 | -0.556 | 0.627 | -0.018 | -0.556 |