Model info
| Transcription factor | ZNF394 (GeneCards) | ||||||||
| Model | ZN394_HUMAN.H11DI.0.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 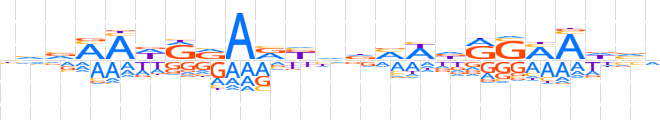 | ||||||||
| LOGO (reverse complement) | 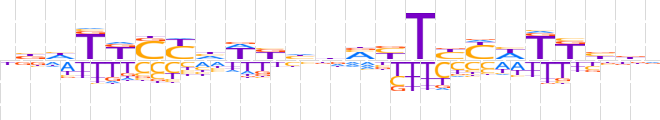 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 23 | ||||||||
| Quality | C | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | dvvMMdRvARbhdvWdRSMAdvh | ||||||||
| Best auROC (human) | 0.912 | ||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | 3 | ||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 390 | ||||||||
| TF family | More than 3 adjacent zinc finger factors {2.3.3} | ||||||||
| TF subfamily | unclassified {2.3.3.0} | ||||||||
| HGNC | HGNC:18832 | ||||||||
| EntrezGene | GeneID:84124 (SSTAR profile) | ||||||||
| UniProt ID | ZN394_HUMAN | ||||||||
| UniProt AC | Q53GI3 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | ZNF394 expression | ||||||||
| ReMap ChIP-seq dataset list | ZNF394 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 43.0 | 7.0 | 20.0 | 5.0 | 57.0 | 6.0 | 2.0 | 8.0 | 33.0 | 15.0 | 9.0 | 19.0 | 33.0 | 74.0 | 38.0 | 9.0 |
| 02 | 56.0 | 52.0 | 48.0 | 10.0 | 45.0 | 7.0 | 49.0 | 1.0 | 18.0 | 14.0 | 30.0 | 7.0 | 25.0 | 2.0 | 11.0 | 3.0 |
| 03 | 96.0 | 25.0 | 22.0 | 1.0 | 59.0 | 9.0 | 3.0 | 4.0 | 92.0 | 20.0 | 21.0 | 5.0 | 14.0 | 1.0 | 6.0 | 0.0 |
| 04 | 224.0 | 20.0 | 8.0 | 9.0 | 45.0 | 2.0 | 1.0 | 7.0 | 30.0 | 12.0 | 8.0 | 2.0 | 2.0 | 5.0 | 1.0 | 2.0 |
| 05 | 95.0 | 13.0 | 45.0 | 148.0 | 17.0 | 3.0 | 1.0 | 18.0 | 7.0 | 0.0 | 4.0 | 7.0 | 8.0 | 0.0 | 2.0 | 10.0 |
| 06 | 22.0 | 9.0 | 85.0 | 11.0 | 4.0 | 5.0 | 4.0 | 3.0 | 12.0 | 5.0 | 26.0 | 9.0 | 26.0 | 5.0 | 147.0 | 5.0 |
| 07 | 35.0 | 6.0 | 22.0 | 1.0 | 16.0 | 3.0 | 4.0 | 1.0 | 38.0 | 31.0 | 172.0 | 21.0 | 8.0 | 3.0 | 12.0 | 5.0 |
| 08 | 92.0 | 3.0 | 2.0 | 0.0 | 39.0 | 0.0 | 0.0 | 4.0 | 200.0 | 3.0 | 4.0 | 3.0 | 27.0 | 1.0 | 0.0 | 0.0 |
| 09 | 167.0 | 61.0 | 126.0 | 4.0 | 5.0 | 2.0 | 0.0 | 0.0 | 0.0 | 2.0 | 3.0 | 1.0 | 2.0 | 1.0 | 4.0 | 0.0 |
| 10 | 19.0 | 21.0 | 32.0 | 102.0 | 7.0 | 7.0 | 4.0 | 48.0 | 19.0 | 19.0 | 22.0 | 73.0 | 1.0 | 1.0 | 0.0 | 3.0 |
| 11 | 25.0 | 5.0 | 4.0 | 12.0 | 19.0 | 13.0 | 4.0 | 12.0 | 33.0 | 7.0 | 4.0 | 14.0 | 60.0 | 74.0 | 26.0 | 66.0 |
| 12 | 36.0 | 8.0 | 76.0 | 17.0 | 19.0 | 6.0 | 61.0 | 13.0 | 2.0 | 11.0 | 24.0 | 1.0 | 18.0 | 18.0 | 27.0 | 41.0 |
| 13 | 37.0 | 18.0 | 10.0 | 10.0 | 25.0 | 11.0 | 5.0 | 2.0 | 129.0 | 32.0 | 17.0 | 10.0 | 22.0 | 33.0 | 14.0 | 3.0 |
| 14 | 173.0 | 21.0 | 12.0 | 7.0 | 31.0 | 7.0 | 1.0 | 55.0 | 20.0 | 5.0 | 3.0 | 18.0 | 17.0 | 0.0 | 3.0 | 5.0 |
| 15 | 64.0 | 10.0 | 53.0 | 114.0 | 9.0 | 4.0 | 4.0 | 16.0 | 7.0 | 3.0 | 4.0 | 5.0 | 19.0 | 1.0 | 53.0 | 12.0 |
| 16 | 37.0 | 1.0 | 57.0 | 4.0 | 6.0 | 3.0 | 4.0 | 5.0 | 58.0 | 2.0 | 45.0 | 9.0 | 20.0 | 0.0 | 123.0 | 4.0 |
| 17 | 16.0 | 15.0 | 90.0 | 0.0 | 3.0 | 0.0 | 1.0 | 2.0 | 17.0 | 32.0 | 174.0 | 6.0 | 0.0 | 4.0 | 15.0 | 3.0 |
| 18 | 19.0 | 11.0 | 4.0 | 2.0 | 26.0 | 13.0 | 1.0 | 11.0 | 195.0 | 46.0 | 13.0 | 26.0 | 7.0 | 2.0 | 2.0 | 0.0 |
| 19 | 215.0 | 14.0 | 16.0 | 2.0 | 57.0 | 7.0 | 4.0 | 4.0 | 6.0 | 9.0 | 2.0 | 3.0 | 28.0 | 5.0 | 1.0 | 5.0 |
| 20 | 82.0 | 26.0 | 37.0 | 161.0 | 8.0 | 8.0 | 2.0 | 17.0 | 11.0 | 3.0 | 3.0 | 6.0 | 6.0 | 2.0 | 1.0 | 5.0 |
| 21 | 53.0 | 24.0 | 18.0 | 12.0 | 8.0 | 26.0 | 1.0 | 4.0 | 12.0 | 22.0 | 5.0 | 4.0 | 20.0 | 108.0 | 49.0 | 12.0 |
| 22 | 34.0 | 19.0 | 9.0 | 31.0 | 114.0 | 29.0 | 4.0 | 33.0 | 26.0 | 24.0 | 10.0 | 13.0 | 17.0 | 7.0 | 3.0 | 5.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.592 | -1.18 | -0.164 | -1.497 | 0.872 | -1.326 | -2.315 | -1.053 | 0.33 | -0.445 | -0.94 | -0.214 | 0.33 | 1.131 | 0.469 | -0.94 |
| 02 | 0.854 | 0.78 | 0.701 | -0.839 | 0.637 | -1.18 | 0.721 | -2.862 | -0.267 | -0.513 | 0.236 | -1.18 | 0.056 | -2.315 | -0.747 | -1.963 |
| 03 | 1.39 | 0.056 | -0.07 | -2.862 | 0.906 | -0.94 | -1.963 | -1.703 | 1.348 | -0.164 | -0.116 | -1.497 | -0.513 | -2.862 | -1.326 | -4.17 |
| 04 | 2.235 | -0.164 | -1.053 | -0.94 | 0.637 | -2.315 | -2.862 | -1.18 | 0.236 | -0.663 | -1.053 | -2.315 | -2.315 | -1.497 | -2.862 | -2.315 |
| 05 | 1.38 | -0.585 | 0.637 | 1.822 | -0.323 | -1.963 | -2.862 | -0.267 | -1.18 | -4.17 | -1.703 | -1.18 | -1.053 | -4.17 | -2.315 | -0.839 |
| 06 | -0.07 | -0.94 | 1.269 | -0.747 | -1.703 | -1.497 | -1.703 | -1.963 | -0.663 | -1.497 | 0.094 | -0.94 | 0.094 | -1.497 | 1.815 | -1.497 |
| 07 | 0.388 | -1.326 | -0.07 | -2.862 | -0.382 | -1.963 | -1.703 | -2.862 | 0.469 | 0.268 | 1.972 | -0.116 | -1.053 | -1.963 | -0.663 | -1.497 |
| 08 | 1.348 | -1.963 | -2.315 | -4.17 | 0.495 | -4.17 | -4.17 | -1.703 | 2.122 | -1.963 | -1.703 | -1.963 | 0.132 | -2.862 | -4.17 | -4.17 |
| 09 | 1.942 | 0.939 | 1.661 | -1.703 | -1.497 | -2.315 | -4.17 | -4.17 | -4.17 | -2.315 | -1.963 | -2.862 | -2.315 | -2.862 | -1.703 | -4.17 |
| 10 | -0.214 | -0.116 | 0.299 | 1.451 | -1.18 | -1.18 | -1.703 | 0.701 | -0.214 | -0.214 | -0.07 | 1.118 | -2.862 | -2.862 | -4.17 | -1.963 |
| 11 | 0.056 | -1.497 | -1.703 | -0.663 | -0.214 | -0.585 | -1.703 | -0.663 | 0.33 | -1.18 | -1.703 | -0.513 | 0.923 | 1.131 | 0.094 | 1.017 |
| 12 | 0.416 | -1.053 | 1.158 | -0.323 | -0.214 | -1.326 | 0.939 | -0.585 | -2.315 | -0.747 | 0.016 | -2.862 | -0.267 | -0.267 | 0.132 | 0.545 |
| 13 | 0.443 | -0.267 | -0.839 | -0.839 | 0.056 | -0.747 | -1.497 | -2.315 | 1.685 | 0.299 | -0.323 | -0.839 | -0.07 | 0.33 | -0.513 | -1.963 |
| 14 | 1.978 | -0.116 | -0.663 | -1.18 | 0.268 | -1.18 | -2.862 | 0.836 | -0.164 | -1.497 | -1.963 | -0.267 | -0.323 | -4.17 | -1.963 | -1.497 |
| 15 | 0.987 | -0.839 | 0.799 | 1.562 | -0.94 | -1.703 | -1.703 | -0.382 | -1.18 | -1.963 | -1.703 | -1.497 | -0.214 | -2.862 | 0.799 | -0.663 |
| 16 | 0.443 | -2.862 | 0.872 | -1.703 | -1.326 | -1.963 | -1.703 | -1.497 | 0.889 | -2.315 | 0.637 | -0.94 | -0.164 | -4.17 | 1.637 | -1.703 |
| 17 | -0.382 | -0.445 | 1.326 | -4.17 | -1.963 | -4.17 | -2.862 | -2.315 | -0.323 | 0.299 | 1.983 | -1.326 | -4.17 | -1.703 | -0.445 | -1.963 |
| 18 | -0.214 | -0.747 | -1.703 | -2.315 | 0.094 | -0.585 | -2.862 | -0.747 | 2.097 | 0.659 | -0.585 | 0.094 | -1.18 | -2.315 | -2.315 | -4.17 |
| 19 | 2.194 | -0.513 | -0.382 | -2.315 | 0.872 | -1.18 | -1.703 | -1.703 | -1.326 | -0.94 | -2.315 | -1.963 | 0.167 | -1.497 | -2.862 | -1.497 |
| 20 | 1.233 | 0.094 | 0.443 | 1.906 | -1.053 | -1.053 | -2.315 | -0.323 | -0.747 | -1.963 | -1.963 | -1.326 | -1.326 | -2.315 | -2.862 | -1.497 |
| 21 | 0.799 | 0.016 | -0.267 | -0.663 | -1.053 | 0.094 | -2.862 | -1.703 | -0.663 | -0.07 | -1.497 | -1.703 | -0.164 | 1.508 | 0.721 | -0.663 |
| 22 | 0.359 | -0.214 | -0.94 | 0.268 | 1.562 | 0.202 | -1.703 | 0.33 | 0.094 | 0.016 | -0.839 | -0.585 | -0.323 | -1.18 | -1.963 | -1.497 |