Model info
| Transcription factor | ZNF449 (GeneCards) | ||||||||
| Model | ZN449_HUMAN.H11DI.0.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 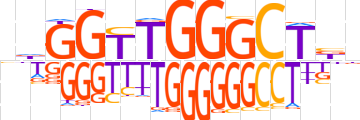 | ||||||||
| LOGO (reverse complement) | 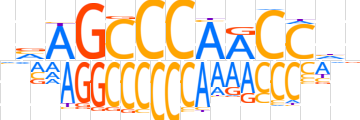 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 13 | ||||||||
| Quality | C | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | ndGGTTGGGCTbn | ||||||||
| Best auROC (human) | 0.941 | ||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | 2 | ||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 392 | ||||||||
| TF family | More than 3 adjacent zinc finger factors {2.3.3} | ||||||||
| TF subfamily | unclassified {2.3.3.0} | ||||||||
| HGNC | HGNC:21039 | ||||||||
| EntrezGene | GeneID:203523 (SSTAR profile) | ||||||||
| UniProt ID | ZN449_HUMAN | ||||||||
| UniProt AC | Q6P9G9 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | ZNF449 expression | ||||||||
| ReMap ChIP-seq dataset list | ZNF449 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 17.0 | 7.0 | 34.0 | 36.0 | 33.0 | 16.0 | 11.0 | 71.0 | 7.0 | 19.0 | 7.0 | 58.0 | 7.0 | 14.0 | 18.0 | 29.0 |
| 02 | 2.0 | 0.0 | 61.0 | 1.0 | 11.0 | 1.0 | 31.0 | 13.0 | 3.0 | 3.0 | 59.0 | 5.0 | 1.0 | 0.0 | 187.0 | 6.0 |
| 03 | 0.0 | 0.0 | 17.0 | 0.0 | 0.0 | 0.0 | 4.0 | 0.0 | 3.0 | 3.0 | 323.0 | 9.0 | 0.0 | 1.0 | 24.0 | 0.0 |
| 04 | 3.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 2.0 | 3.0 | 88.0 | 0.0 | 277.0 | 0.0 | 3.0 | 0.0 | 6.0 |
| 05 | 1.0 | 1.0 | 0.0 | 4.0 | 1.0 | 1.0 | 11.0 | 80.0 | 0.0 | 0.0 | 0.0 | 0.0 | 12.0 | 4.0 | 9.0 | 260.0 |
| 06 | 0.0 | 0.0 | 14.0 | 0.0 | 0.0 | 0.0 | 6.0 | 0.0 | 0.0 | 0.0 | 20.0 | 0.0 | 0.0 | 0.0 | 344.0 | 0.0 |
| 07 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 384.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 08 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.0 | 27.0 | 349.0 | 5.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 09 | 0.0 | 3.0 | 0.0 | 0.0 | 0.0 | 27.0 | 0.0 | 0.0 | 1.0 | 344.0 | 3.0 | 1.0 | 0.0 | 5.0 | 0.0 | 0.0 |
| 10 | 0.0 | 0.0 | 0.0 | 1.0 | 29.0 | 6.0 | 15.0 | 329.0 | 0.0 | 1.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 1.0 |
| 11 | 1.0 | 10.0 | 5.0 | 13.0 | 1.0 | 2.0 | 0.0 | 4.0 | 1.0 | 7.0 | 3.0 | 4.0 | 3.0 | 75.0 | 104.0 | 151.0 |
| 12 | 2.0 | 1.0 | 3.0 | 0.0 | 31.0 | 21.0 | 10.0 | 32.0 | 40.0 | 12.0 | 37.0 | 23.0 | 14.0 | 44.0 | 52.0 | 62.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.339 | -1.196 | 0.344 | 0.4 | 0.314 | -0.398 | -0.762 | 1.074 | -1.196 | -0.23 | -1.196 | 0.873 | -1.196 | -0.528 | -0.283 | 0.187 |
| 02 | -2.33 | -4.183 | 0.924 | -2.877 | -0.762 | -2.877 | 0.252 | -0.6 | -1.978 | -1.978 | 0.89 | -1.512 | -2.877 | -4.183 | 2.04 | -1.342 |
| 03 | -4.183 | -4.183 | -0.339 | -4.183 | -4.183 | -4.183 | -1.718 | -4.183 | -1.978 | -1.978 | 2.585 | -0.956 | -4.183 | -2.877 | 0.0 | -4.183 |
| 04 | -1.978 | -4.183 | -4.183 | -4.183 | -4.183 | -2.33 | -4.183 | -2.33 | -1.978 | 1.288 | -4.183 | 2.432 | -4.183 | -1.978 | -4.183 | -1.342 |
| 05 | -2.877 | -2.877 | -4.183 | -1.718 | -2.877 | -2.877 | -0.762 | 1.193 | -4.183 | -4.183 | -4.183 | -4.183 | -0.678 | -1.718 | -0.956 | 2.369 |
| 06 | -4.183 | -4.183 | -0.528 | -4.183 | -4.183 | -4.183 | -1.342 | -4.183 | -4.183 | -4.183 | -0.179 | -4.183 | -4.183 | -4.183 | 2.648 | -4.183 |
| 07 | -4.183 | -4.183 | -4.183 | -4.183 | -4.183 | -4.183 | -4.183 | -4.183 | -4.183 | -4.183 | 2.758 | -4.183 | -4.183 | -4.183 | -4.183 | -4.183 |
| 08 | -4.183 | -4.183 | -4.183 | -4.183 | -4.183 | -4.183 | -4.183 | -4.183 | -1.978 | 0.116 | 2.663 | -1.512 | -4.183 | -4.183 | -4.183 | -4.183 |
| 09 | -4.183 | -1.978 | -4.183 | -4.183 | -4.183 | 0.116 | -4.183 | -4.183 | -2.877 | 2.648 | -1.978 | -2.877 | -4.183 | -1.512 | -4.183 | -4.183 |
| 10 | -4.183 | -4.183 | -4.183 | -2.877 | 0.187 | -1.342 | -0.461 | 2.604 | -4.183 | -2.877 | -4.183 | -2.33 | -4.183 | -4.183 | -4.183 | -2.877 |
| 11 | -2.877 | -0.854 | -1.512 | -0.6 | -2.877 | -2.33 | -4.183 | -1.718 | -2.877 | -1.196 | -1.978 | -1.718 | -1.978 | 1.129 | 1.455 | 1.826 |
| 12 | -2.33 | -2.877 | -1.978 | -4.183 | 0.252 | -0.131 | -0.854 | 0.284 | 0.505 | -0.678 | 0.427 | -0.042 | -0.528 | 0.599 | 0.765 | 0.94 |