Model info
| Transcription factor | ZNF554 (GeneCards) | ||||||||
| Model | ZN554_HUMAN.H11MO.0.C | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 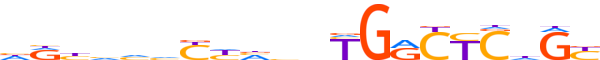 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 20 | ||||||||
| Quality | C | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | RCdGRGYCRnbbRRdddvYh | ||||||||
| Best auROC (human) | 0.853 | ||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | 2 | ||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 501 | ||||||||
| TF family | More than 3 adjacent zinc finger factors {2.3.3} | ||||||||
| TF subfamily | unclassified {2.3.3.0} | ||||||||
| HGNC | HGNC:26629 | ||||||||
| EntrezGene | GeneID:115196 (SSTAR profile) | ||||||||
| UniProt ID | ZN554_HUMAN | ||||||||
| UniProt AC | Q86TJ5 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | ZNF554 expression | ||||||||
| ReMap ChIP-seq dataset list | ZNF554 datasets | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 120.0 | 47.0 | 300.0 | 33.0 |
| 02 | 30.0 | 412.0 | 3.0 | 55.0 |
| 03 | 172.0 | 57.0 | 77.0 | 194.0 |
| 04 | 22.0 | 32.0 | 423.0 | 23.0 |
| 05 | 343.0 | 5.0 | 142.0 | 10.0 |
| 06 | 71.0 | 19.0 | 408.0 | 2.0 |
| 07 | 8.0 | 285.0 | 23.0 | 184.0 |
| 08 | 0.0 | 493.0 | 0.0 | 7.0 |
| 09 | 389.0 | 30.0 | 45.0 | 36.0 |
| 10 | 87.0 | 136.0 | 120.0 | 157.0 |
| 11 | 89.0 | 101.0 | 216.0 | 94.0 |
| 12 | 60.0 | 80.0 | 84.0 | 276.0 |
| 13 | 207.0 | 20.0 | 218.0 | 55.0 |
| 14 | 70.0 | 59.0 | 351.0 | 20.0 |
| 15 | 109.0 | 49.0 | 188.0 | 154.0 |
| 16 | 69.0 | 60.0 | 228.0 | 143.0 |
| 17 | 80.0 | 62.0 | 137.0 | 221.0 |
| 18 | 191.0 | 53.0 | 212.0 | 44.0 |
| 19 | 63.0 | 327.0 | 45.0 | 65.0 |
| 20 | 92.0 | 77.0 | 72.0 | 259.0 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.04 | -0.958 | 0.868 | -1.298 |
| 02 | -1.389 | 1.184 | -3.325 | -0.805 |
| 03 | 0.316 | -0.771 | -0.477 | 0.435 |
| 04 | -1.681 | -1.328 | 1.21 | -1.64 |
| 05 | 1.002 | -2.961 | 0.126 | -2.394 |
| 06 | -0.556 | -1.818 | 1.174 | -3.573 |
| 07 | -2.584 | 0.817 | -1.64 | 0.383 |
| 08 | -4.4 | 1.363 | -4.4 | -2.694 |
| 09 | 1.127 | -1.389 | -1.0 | -1.215 |
| 10 | -0.357 | 0.083 | -0.04 | 0.225 |
| 11 | -0.335 | -0.21 | 0.542 | -0.281 |
| 12 | -0.721 | -0.439 | -0.392 | 0.785 |
| 13 | 0.5 | -1.77 | 0.551 | -0.805 |
| 14 | -0.57 | -0.737 | 1.025 | -1.77 |
| 15 | -0.135 | -0.918 | 0.404 | 0.206 |
| 16 | -0.584 | -0.721 | 0.595 | 0.133 |
| 17 | -0.439 | -0.689 | 0.091 | 0.565 |
| 18 | 0.42 | -0.841 | 0.523 | -1.022 |
| 19 | -0.673 | 0.954 | -1.0 | -0.643 |
| 20 | -0.302 | -0.477 | -0.543 | 0.722 |