Model info
| Transcription factor | ZNF586 (GeneCards) | ||||||||
| Model | ZN586_HUMAN.H11MO.0.C | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO | 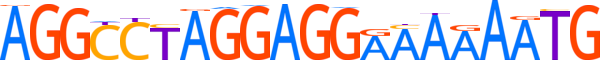 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 20 | ||||||||
| Quality | C | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | AGGCCTAGGAGGAAAAAATG | ||||||||
| Best auROC (human) | 1.0 | ||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | 4 | ||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 472 | ||||||||
| TF family | More than 3 adjacent zinc finger factors {2.3.3} | ||||||||
| TF subfamily | unclassified {2.3.3.0} | ||||||||
| HGNC | HGNC:25949 | ||||||||
| EntrezGene | GeneID:54807 (SSTAR profile) | ||||||||
| UniProt ID | ZN586_HUMAN | ||||||||
| UniProt AC | Q9NXT0 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | ZNF586 expression | ||||||||
| ReMap ChIP-seq dataset list | ZNF586 datasets | ||||||||
| Motifs in JASPAR |
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 462.0 | 0.0 | 9.0 | 1.0 |
| 02 | 6.0 | 0.0 | 465.0 | 1.0 |
| 03 | 7.0 | 4.0 | 458.0 | 3.0 |
| 04 | 12.0 | 414.0 | 5.0 | 41.0 |
| 05 | 6.0 | 419.0 | 3.0 | 44.0 |
| 06 | 7.0 | 56.0 | 8.0 | 401.0 |
| 07 | 451.0 | 2.0 | 16.0 | 3.0 |
| 08 | 19.0 | 0.0 | 453.0 | 0.0 |
| 09 | 9.0 | 1.0 | 462.0 | 0.0 |
| 10 | 466.0 | 0.0 | 4.0 | 2.0 |
| 11 | 3.0 | 0.0 | 466.0 | 3.0 |
| 12 | 10.0 | 0.0 | 457.0 | 5.0 |
| 13 | 353.0 | 6.0 | 109.0 | 4.0 |
| 14 | 399.0 | 56.0 | 4.0 | 13.0 |
| 15 | 443.0 | 3.0 | 4.0 | 22.0 |
| 16 | 384.0 | 4.0 | 74.0 | 10.0 |
| 17 | 472.0 | 0.0 | 0.0 | 0.0 |
| 18 | 423.0 | 2.0 | 47.0 | 0.0 |
| 19 | 4.0 | 14.0 | 3.0 | 451.0 |
| 20 | 7.0 | 0.0 | 464.0 | 1.0 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 1.355 | -4.352 | -2.429 | -3.852 |
| 02 | -2.764 | -4.352 | 1.362 | -3.852 |
| 03 | -2.639 | -3.072 | 1.347 | -3.271 |
| 04 | -2.178 | 1.246 | -2.906 | -1.033 |
| 05 | -2.764 | 1.258 | -3.271 | -0.965 |
| 06 | -2.639 | -0.731 | -2.528 | 1.214 |
| 07 | 1.331 | -3.52 | -1.919 | -3.271 |
| 08 | -1.761 | -4.352 | 1.336 | -4.352 |
| 09 | -2.429 | -3.852 | 1.355 | -4.352 |
| 10 | 1.364 | -4.352 | -3.072 | -3.52 |
| 11 | -3.271 | -4.352 | 1.364 | -3.271 |
| 12 | -2.338 | -4.352 | 1.344 | -2.906 |
| 13 | 1.087 | -2.764 | -0.078 | -3.072 |
| 14 | 1.209 | -0.731 | -3.072 | -2.107 |
| 15 | 1.313 | -3.271 | -3.072 | -1.625 |
| 16 | 1.171 | -3.072 | -0.459 | -2.338 |
| 17 | 1.377 | -4.352 | -4.352 | -4.352 |
| 18 | 1.267 | -3.52 | -0.901 | -4.352 |
| 19 | -3.072 | -2.04 | -3.271 | 1.331 |
| 20 | -2.639 | -4.352 | 1.36 | -3.852 |