Model info
| Transcription factor | ZNF708 (GeneCards) | ||||||||
| Model | ZN708_HUMAN.H11DI.0.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 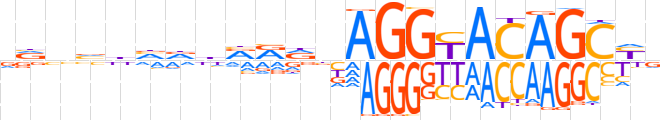 | ||||||||
| LOGO (reverse complement) | 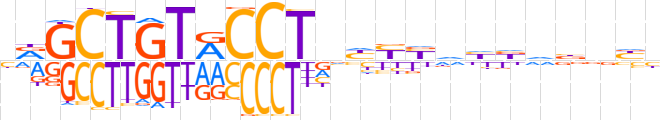 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 23 | ||||||||
| Quality | C | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | vvnvhvddRRdnAGGYACAGCYv | ||||||||
| Best auROC (human) | 0.856 | ||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | 3 | ||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 312 | ||||||||
| TF family | More than 3 adjacent zinc finger factors {2.3.3} | ||||||||
| TF subfamily | unclassified {2.3.3.0} | ||||||||
| HGNC | HGNC:12945 | ||||||||
| EntrezGene | GeneID:7562 (SSTAR profile) | ||||||||
| UniProt ID | ZN708_HUMAN | ||||||||
| UniProt AC | P17019 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | ZNF708 expression | ||||||||
| ReMap ChIP-seq dataset list | ZNF708 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 19.0 | 3.0 | 48.0 | 9.0 | 19.0 | 16.0 | 3.0 | 13.0 | 10.0 | 11.0 | 110.0 | 8.0 | 10.0 | 10.0 | 19.0 | 4.0 |
| 02 | 19.0 | 12.0 | 18.0 | 9.0 | 11.0 | 7.0 | 3.0 | 19.0 | 35.0 | 92.0 | 34.0 | 19.0 | 7.0 | 5.0 | 17.0 | 5.0 |
| 03 | 20.0 | 24.0 | 21.0 | 7.0 | 17.0 | 88.0 | 0.0 | 11.0 | 22.0 | 19.0 | 19.0 | 12.0 | 11.0 | 17.0 | 18.0 | 6.0 |
| 04 | 22.0 | 12.0 | 19.0 | 17.0 | 19.0 | 14.0 | 7.0 | 108.0 | 16.0 | 16.0 | 18.0 | 8.0 | 3.0 | 14.0 | 10.0 | 9.0 |
| 05 | 29.0 | 11.0 | 12.0 | 8.0 | 19.0 | 21.0 | 5.0 | 11.0 | 18.0 | 16.0 | 11.0 | 9.0 | 106.0 | 13.0 | 15.0 | 8.0 |
| 06 | 123.0 | 8.0 | 34.0 | 7.0 | 25.0 | 10.0 | 4.0 | 22.0 | 11.0 | 7.0 | 18.0 | 7.0 | 5.0 | 6.0 | 17.0 | 8.0 |
| 07 | 24.0 | 19.0 | 24.0 | 97.0 | 13.0 | 8.0 | 0.0 | 10.0 | 12.0 | 17.0 | 20.0 | 24.0 | 14.0 | 2.0 | 15.0 | 13.0 |
| 08 | 35.0 | 10.0 | 14.0 | 4.0 | 31.0 | 7.0 | 0.0 | 8.0 | 21.0 | 11.0 | 17.0 | 10.0 | 102.0 | 12.0 | 22.0 | 8.0 |
| 09 | 112.0 | 11.0 | 60.0 | 6.0 | 34.0 | 2.0 | 1.0 | 3.0 | 21.0 | 12.0 | 19.0 | 1.0 | 8.0 | 2.0 | 18.0 | 2.0 |
| 10 | 55.0 | 3.0 | 104.0 | 13.0 | 3.0 | 5.0 | 4.0 | 15.0 | 20.0 | 6.0 | 49.0 | 23.0 | 0.0 | 1.0 | 9.0 | 2.0 |
| 11 | 13.0 | 28.0 | 13.0 | 24.0 | 8.0 | 2.0 | 1.0 | 4.0 | 13.0 | 53.0 | 47.0 | 53.0 | 15.0 | 11.0 | 18.0 | 9.0 |
| 12 | 44.0 | 0.0 | 2.0 | 3.0 | 92.0 | 0.0 | 2.0 | 0.0 | 76.0 | 0.0 | 2.0 | 1.0 | 85.0 | 0.0 | 5.0 | 0.0 |
| 13 | 0.0 | 0.0 | 296.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 10.0 | 1.0 | 0.0 | 1.0 | 3.0 | 0.0 |
| 14 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 1.0 | 9.0 | 297.0 | 2.0 | 0.0 | 0.0 | 2.0 | 0.0 |
| 15 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 9.0 | 0.0 | 0.0 | 7.0 | 113.0 | 0.0 | 180.0 | 0.0 | 1.0 | 0.0 | 1.0 |
| 16 | 6.0 | 0.0 | 1.0 | 0.0 | 120.0 | 2.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 177.0 | 1.0 | 2.0 | 1.0 |
| 17 | 0.0 | 264.0 | 3.0 | 36.0 | 0.0 | 3.0 | 0.0 | 0.0 | 1.0 | 4.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 |
| 18 | 1.0 | 0.0 | 0.0 | 0.0 | 256.0 | 0.0 | 15.0 | 1.0 | 3.0 | 0.0 | 0.0 | 0.0 | 27.0 | 0.0 | 9.0 | 0.0 |
| 19 | 3.0 | 2.0 | 280.0 | 2.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.0 | 0.0 | 20.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 |
| 20 | 2.0 | 4.0 | 0.0 | 1.0 | 0.0 | 3.0 | 0.0 | 0.0 | 16.0 | 266.0 | 6.0 | 12.0 | 0.0 | 2.0 | 0.0 | 0.0 |
| 21 | 9.0 | 2.0 | 1.0 | 6.0 | 48.0 | 67.0 | 3.0 | 157.0 | 2.0 | 3.0 | 1.0 | 0.0 | 2.0 | 6.0 | 0.0 | 5.0 |
| 22 | 17.0 | 9.0 | 20.0 | 15.0 | 33.0 | 20.0 | 4.0 | 21.0 | 0.0 | 4.0 | 1.0 | 0.0 | 15.0 | 22.0 | 114.0 | 17.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.025 | -1.777 | 0.89 | -0.752 | -0.025 | -0.194 | -1.777 | -0.396 | -0.651 | -0.559 | 1.715 | -0.865 | -0.651 | -0.651 | -0.025 | -1.516 |
| 02 | -0.025 | -0.474 | -0.079 | -0.752 | -0.559 | -0.993 | -1.777 | -0.025 | 0.577 | 1.537 | 0.548 | -0.025 | -0.993 | -1.31 | -0.135 | -1.31 |
| 03 | 0.025 | 0.204 | 0.073 | -0.993 | -0.135 | 1.493 | -4.013 | -0.559 | 0.119 | -0.025 | -0.025 | -0.474 | -0.559 | -0.135 | -0.079 | -1.139 |
| 04 | 0.119 | -0.474 | -0.025 | -0.135 | -0.025 | -0.324 | -0.993 | 1.697 | -0.194 | -0.194 | -0.079 | -0.865 | -1.777 | -0.324 | -0.651 | -0.752 |
| 05 | 0.391 | -0.559 | -0.474 | -0.865 | -0.025 | 0.073 | -1.31 | -0.559 | -0.079 | -0.194 | -0.559 | -0.752 | 1.678 | -0.396 | -0.257 | -0.865 |
| 06 | 1.826 | -0.865 | 0.548 | -0.993 | 0.244 | -0.651 | -1.516 | 0.119 | -0.559 | -0.993 | -0.079 | -0.993 | -1.31 | -1.139 | -0.135 | -0.865 |
| 07 | 0.204 | -0.025 | 0.204 | 1.59 | -0.396 | -0.865 | -4.013 | -0.651 | -0.474 | -0.135 | 0.025 | 0.204 | -0.324 | -2.13 | -0.257 | -0.396 |
| 08 | 0.577 | -0.651 | -0.324 | -1.516 | 0.457 | -0.993 | -4.013 | -0.865 | 0.073 | -0.559 | -0.135 | -0.651 | 1.64 | -0.474 | 0.119 | -0.865 |
| 09 | 1.733 | -0.559 | 1.112 | -1.139 | 0.548 | -2.13 | -2.682 | -1.777 | 0.073 | -0.474 | -0.025 | -2.682 | -0.865 | -2.13 | -0.079 | -2.13 |
| 10 | 1.025 | -1.777 | 1.659 | -0.396 | -1.777 | -1.31 | -1.516 | -0.257 | 0.025 | -1.139 | 0.91 | 0.162 | -4.013 | -2.682 | -0.752 | -2.13 |
| 11 | -0.396 | 0.356 | -0.396 | 0.204 | -0.865 | -2.13 | -2.682 | -1.516 | -0.396 | 0.988 | 0.869 | 0.988 | -0.257 | -0.559 | -0.079 | -0.752 |
| 12 | 0.804 | -4.013 | -2.13 | -1.777 | 1.537 | -4.013 | -2.13 | -4.013 | 1.347 | -4.013 | -2.13 | -2.682 | 1.458 | -4.013 | -1.31 | -4.013 |
| 13 | -4.013 | -4.013 | 2.703 | -2.682 | -4.013 | -4.013 | -4.013 | -4.013 | -4.013 | -4.013 | -0.651 | -2.682 | -4.013 | -2.682 | -1.777 | -4.013 |
| 14 | -4.013 | -4.013 | -4.013 | -4.013 | -4.013 | -4.013 | -2.682 | -4.013 | -2.682 | -0.752 | 2.706 | -2.13 | -4.013 | -4.013 | -2.13 | -4.013 |
| 15 | -4.013 | -2.682 | -4.013 | -4.013 | -4.013 | -0.752 | -4.013 | -4.013 | -0.993 | 1.742 | -4.013 | 2.206 | -4.013 | -2.682 | -4.013 | -2.682 |
| 16 | -1.139 | -4.013 | -2.682 | -4.013 | 1.802 | -2.13 | -2.13 | -4.013 | -4.013 | -4.013 | -4.013 | -4.013 | 2.19 | -2.682 | -2.13 | -2.682 |
| 17 | -4.013 | 2.589 | -1.777 | 0.605 | -4.013 | -1.777 | -4.013 | -4.013 | -2.682 | -1.516 | -4.013 | -4.013 | -4.013 | -2.682 | -4.013 | -4.013 |
| 18 | -2.682 | -4.013 | -4.013 | -4.013 | 2.558 | -4.013 | -0.257 | -2.682 | -1.777 | -4.013 | -4.013 | -4.013 | 0.32 | -4.013 | -0.752 | -4.013 |
| 19 | -1.777 | -2.13 | 2.647 | -2.13 | -4.013 | -4.013 | -4.013 | -4.013 | -1.516 | -4.013 | 0.025 | -4.013 | -4.013 | -2.682 | -4.013 | -4.013 |
| 20 | -2.13 | -1.516 | -4.013 | -2.682 | -4.013 | -1.777 | -4.013 | -4.013 | -0.194 | 2.596 | -1.139 | -0.474 | -4.013 | -2.13 | -4.013 | -4.013 |
| 21 | -0.752 | -2.13 | -2.682 | -1.139 | 0.89 | 1.221 | -1.777 | 2.07 | -2.13 | -1.777 | -2.682 | -4.013 | -2.13 | -1.139 | -4.013 | -1.31 |
| 22 | -0.135 | -0.752 | 0.025 | -0.257 | 0.519 | 0.025 | -1.516 | 0.073 | -4.013 | -1.516 | -2.682 | -4.013 | -0.257 | 0.119 | 1.751 | -0.135 |