Model info
| Transcription factor | ZNF770 (GeneCards) | ||||||||
| Model | ZN770_HUMAN.H11DI.0.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 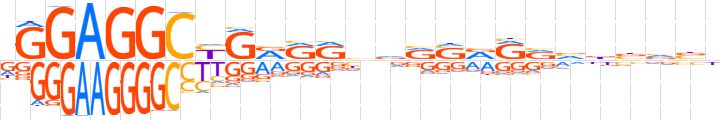 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 25 | ||||||||
| Quality | C | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | dGGAGGCYGRRRbvRSRRvvbbvbb | ||||||||
| Best auROC (human) | 0.981 | ||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | 3 | ||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 476 | ||||||||
| TF family | Factors with multiple dispersed zinc fingers {2.3.4} | ||||||||
| TF subfamily | unclassified {2.3.4.0} | ||||||||
| HGNC | HGNC:26061 | ||||||||
| EntrezGene | GeneID:54989 (SSTAR profile) | ||||||||
| UniProt ID | ZN770_HUMAN | ||||||||
| UniProt AC | Q6IQ21 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | ZNF770 expression | ||||||||
| ReMap ChIP-seq dataset list | ZNF770 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 9.0 | 0.0 | 94.0 | 2.0 | 9.0 | 0.0 | 50.0 | 4.0 | 20.0 | 1.0 | 191.0 | 6.0 | 18.0 | 1.0 | 68.0 | 1.0 |
| 02 | 1.0 | 0.0 | 55.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 10.0 | 2.0 | 390.0 | 1.0 | 0.0 | 0.0 | 13.0 | 0.0 |
| 03 | 11.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 | 0.0 | 0.0 | 454.0 | 0.0 | 1.0 | 5.0 | 1.0 | 0.0 | 0.0 | 0.0 |
| 04 | 7.0 | 4.0 | 456.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 5.0 | 0.0 |
| 05 | 0.0 | 0.0 | 7.0 | 1.0 | 0.0 | 0.0 | 4.0 | 0.0 | 5.0 | 1.0 | 451.0 | 4.0 | 0.0 | 0.0 | 1.0 | 0.0 |
| 06 | 1.0 | 4.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 13.0 | 438.0 | 6.0 | 6.0 | 0.0 | 5.0 | 0.0 | 0.0 |
| 07 | 2.0 | 3.0 | 4.0 | 5.0 | 30.0 | 136.0 | 13.0 | 269.0 | 2.0 | 1.0 | 3.0 | 0.0 | 0.0 | 2.0 | 4.0 | 0.0 |
| 08 | 1.0 | 1.0 | 32.0 | 0.0 | 31.0 | 23.0 | 86.0 | 2.0 | 0.0 | 2.0 | 22.0 | 0.0 | 8.0 | 9.0 | 249.0 | 8.0 |
| 09 | 14.0 | 2.0 | 22.0 | 2.0 | 17.0 | 4.0 | 11.0 | 3.0 | 234.0 | 37.0 | 104.0 | 14.0 | 4.0 | 3.0 | 1.0 | 2.0 |
| 10 | 28.0 | 16.0 | 222.0 | 3.0 | 10.0 | 10.0 | 19.0 | 7.0 | 32.0 | 27.0 | 72.0 | 7.0 | 3.0 | 4.0 | 11.0 | 3.0 |
| 11 | 19.0 | 6.0 | 47.0 | 1.0 | 13.0 | 10.0 | 24.0 | 10.0 | 76.0 | 13.0 | 224.0 | 11.0 | 1.0 | 1.0 | 16.0 | 2.0 |
| 12 | 27.0 | 21.0 | 47.0 | 14.0 | 5.0 | 14.0 | 3.0 | 8.0 | 41.0 | 138.0 | 54.0 | 78.0 | 2.0 | 14.0 | 7.0 | 1.0 |
| 13 | 17.0 | 7.0 | 41.0 | 10.0 | 77.0 | 30.0 | 51.0 | 29.0 | 34.0 | 24.0 | 41.0 | 12.0 | 12.0 | 12.0 | 69.0 | 8.0 |
| 14 | 12.0 | 10.0 | 105.0 | 13.0 | 22.0 | 15.0 | 27.0 | 9.0 | 39.0 | 31.0 | 128.0 | 4.0 | 4.0 | 14.0 | 34.0 | 7.0 |
| 15 | 18.0 | 3.0 | 55.0 | 1.0 | 13.0 | 25.0 | 15.0 | 17.0 | 31.0 | 34.0 | 220.0 | 9.0 | 1.0 | 8.0 | 18.0 | 6.0 |
| 16 | 30.0 | 7.0 | 22.0 | 4.0 | 22.0 | 16.0 | 18.0 | 14.0 | 211.0 | 19.0 | 65.0 | 13.0 | 3.0 | 3.0 | 22.0 | 5.0 |
| 17 | 16.0 | 14.0 | 231.0 | 5.0 | 10.0 | 13.0 | 14.0 | 8.0 | 35.0 | 23.0 | 61.0 | 8.0 | 2.0 | 5.0 | 27.0 | 2.0 |
| 18 | 12.0 | 11.0 | 38.0 | 2.0 | 7.0 | 27.0 | 6.0 | 15.0 | 62.0 | 36.0 | 216.0 | 19.0 | 3.0 | 8.0 | 12.0 | 0.0 |
| 19 | 44.0 | 12.0 | 24.0 | 4.0 | 10.0 | 40.0 | 19.0 | 13.0 | 160.0 | 67.0 | 28.0 | 17.0 | 4.0 | 14.0 | 14.0 | 4.0 |
| 20 | 14.0 | 24.0 | 43.0 | 137.0 | 19.0 | 30.0 | 36.0 | 48.0 | 26.0 | 8.0 | 39.0 | 12.0 | 4.0 | 6.0 | 14.0 | 14.0 |
| 21 | 8.0 | 11.0 | 40.0 | 4.0 | 14.0 | 17.0 | 10.0 | 27.0 | 14.0 | 34.0 | 69.0 | 15.0 | 3.0 | 141.0 | 33.0 | 34.0 |
| 22 | 11.0 | 4.0 | 22.0 | 2.0 | 74.0 | 34.0 | 77.0 | 18.0 | 45.0 | 45.0 | 52.0 | 10.0 | 11.0 | 14.0 | 44.0 | 11.0 |
| 23 | 14.0 | 69.0 | 49.0 | 9.0 | 13.0 | 46.0 | 6.0 | 32.0 | 29.0 | 107.0 | 48.0 | 11.0 | 0.0 | 16.0 | 14.0 | 11.0 |
| 24 | 11.0 | 13.0 | 27.0 | 5.0 | 18.0 | 34.0 | 28.0 | 158.0 | 25.0 | 35.0 | 46.0 | 11.0 | 4.0 | 12.0 | 28.0 | 19.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -1.162 | -4.356 | 1.146 | -2.532 | -1.162 | -4.356 | 0.518 | -1.923 | -0.387 | -3.076 | 1.853 | -1.548 | -0.49 | -3.076 | 0.824 | -3.076 |
| 02 | -3.076 | -4.356 | 0.613 | -4.356 | -4.356 | -4.356 | -2.532 | -4.356 | -1.061 | -2.532 | 2.566 | -3.076 | -4.356 | -4.356 | -0.807 | -4.356 |
| 03 | -0.969 | -4.356 | -4.356 | -4.356 | -2.532 | -4.356 | -4.356 | -4.356 | 2.717 | -4.356 | -3.076 | -1.718 | -3.076 | -4.356 | -4.356 | -4.356 |
| 04 | -1.402 | -1.923 | 2.722 | -3.076 | -4.356 | -4.356 | -4.356 | -4.356 | -3.076 | -4.356 | -4.356 | -4.356 | -4.356 | -4.356 | -1.718 | -4.356 |
| 05 | -4.356 | -4.356 | -1.402 | -3.076 | -4.356 | -4.356 | -1.923 | -4.356 | -1.718 | -3.076 | 2.711 | -1.923 | -4.356 | -4.356 | -3.076 | -4.356 |
| 06 | -3.076 | -1.923 | -4.356 | -4.356 | -4.356 | -3.076 | -4.356 | -4.356 | -0.807 | 2.682 | -1.548 | -1.548 | -4.356 | -1.718 | -4.356 | -4.356 |
| 07 | -2.532 | -2.182 | -1.923 | -1.718 | 0.012 | 1.514 | -0.807 | 2.195 | -2.532 | -3.076 | -2.182 | -4.356 | -4.356 | -2.532 | -1.923 | -4.356 |
| 08 | -3.076 | -3.076 | 0.076 | -4.356 | 0.045 | -0.249 | 1.057 | -2.532 | -4.356 | -2.532 | -0.293 | -4.356 | -1.275 | -1.162 | 2.117 | -1.275 |
| 09 | -0.735 | -2.532 | -0.293 | -2.532 | -0.546 | -1.923 | -0.969 | -2.182 | 2.055 | 0.22 | 1.247 | -0.735 | -1.923 | -2.182 | -3.076 | -2.532 |
| 10 | -0.056 | -0.605 | 2.003 | -2.182 | -1.061 | -1.061 | -0.437 | -1.402 | 0.076 | -0.092 | 0.88 | -1.402 | -2.182 | -1.923 | -0.969 | -2.182 |
| 11 | -0.437 | -1.548 | 0.457 | -3.076 | -0.807 | -1.061 | -0.208 | -1.061 | 0.934 | -0.807 | 2.012 | -0.969 | -3.076 | -3.076 | -0.605 | -2.532 |
| 12 | -0.092 | -0.339 | 0.457 | -0.735 | -1.718 | -0.735 | -2.182 | -1.275 | 0.321 | 1.529 | 0.595 | 0.96 | -2.532 | -0.735 | -1.402 | -3.076 |
| 13 | -0.546 | -1.402 | 0.321 | -1.061 | 0.947 | 0.012 | 0.538 | -0.021 | 0.136 | -0.208 | 0.321 | -0.885 | -0.885 | -0.885 | 0.838 | -1.275 |
| 14 | -0.885 | -1.061 | 1.256 | -0.807 | -0.293 | -0.668 | -0.092 | -1.162 | 0.272 | 0.045 | 1.454 | -1.923 | -1.923 | -0.735 | 0.136 | -1.402 |
| 15 | -0.49 | -2.182 | 0.613 | -3.076 | -0.807 | -0.167 | -0.668 | -0.546 | 0.045 | 0.136 | 1.994 | -1.162 | -3.076 | -1.275 | -0.49 | -1.548 |
| 16 | 0.012 | -1.402 | -0.293 | -1.923 | -0.293 | -0.605 | -0.49 | -0.735 | 1.952 | -0.437 | 0.779 | -0.807 | -2.182 | -2.182 | -0.293 | -1.718 |
| 17 | -0.605 | -0.735 | 2.043 | -1.718 | -1.061 | -0.807 | -0.735 | -1.275 | 0.165 | -0.249 | 0.716 | -1.275 | -2.532 | -1.718 | -0.092 | -2.532 |
| 18 | -0.885 | -0.969 | 0.246 | -2.532 | -1.402 | -0.092 | -1.548 | -0.668 | 0.732 | 0.193 | 1.976 | -0.437 | -2.182 | -1.275 | -0.885 | -4.356 |
| 19 | 0.391 | -0.885 | -0.208 | -1.923 | -1.061 | 0.297 | -0.437 | -0.807 | 1.676 | 0.809 | -0.056 | -0.546 | -1.923 | -0.735 | -0.735 | -1.923 |
| 20 | -0.735 | -0.208 | 0.369 | 1.521 | -0.437 | 0.012 | 0.193 | 0.478 | -0.129 | -1.275 | 0.272 | -0.885 | -1.923 | -1.548 | -0.735 | -0.735 |
| 21 | -1.275 | -0.969 | 0.297 | -1.923 | -0.735 | -0.546 | -1.061 | -0.092 | -0.735 | 0.136 | 0.838 | -0.668 | -2.182 | 1.55 | 0.107 | 0.136 |
| 22 | -0.969 | -1.923 | -0.293 | -2.532 | 0.908 | 0.136 | 0.947 | -0.49 | 0.414 | 0.414 | 0.557 | -1.061 | -0.969 | -0.735 | 0.391 | -0.969 |
| 23 | -0.735 | 0.838 | 0.498 | -1.162 | -0.807 | 0.435 | -1.548 | 0.076 | -0.021 | 1.275 | 0.478 | -0.969 | -4.356 | -0.605 | -0.735 | -0.969 |
| 24 | -0.969 | -0.807 | -0.092 | -1.718 | -0.49 | 0.136 | -0.056 | 1.663 | -0.167 | 0.165 | 0.435 | -0.969 | -1.923 | -0.885 | -0.056 | -0.437 |