Model info
| Transcription factor | ZNF816 (GeneCards) | ||||||||
| Model | ZN816_HUMAN.H11DI.0.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 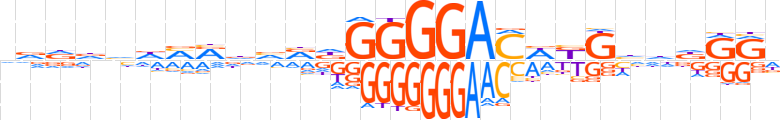 | ||||||||
| LOGO (reverse complement) | 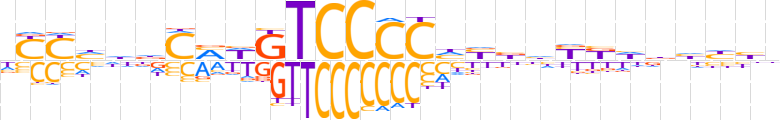 | ||||||||
| Data source | ChIP-Seq | ||||||||
| Model release | HOCOMOCOv11 | ||||||||
| Model length | 27 | ||||||||
| Quality | C | ||||||||
| Motif rank | 0 | ||||||||
| Consensus | nvddhdMRdvvdGGGGACWKGhhdRRd | ||||||||
| Best auROC (human) | 0.968 | ||||||||
| Best auROC (mouse) | |||||||||
| Peak sets in benchmark (human) | 4 | ||||||||
| Peak sets in benchmark (mouse) | |||||||||
| Aligned words | 475 | ||||||||
| TF family | More than 3 adjacent zinc finger factors {2.3.3} | ||||||||
| TF subfamily | ZNF816A-like factors {2.3.3.73} | ||||||||
| HGNC | HGNC:26995 | ||||||||
| EntrezGene | GeneID:125893 (SSTAR profile) | ||||||||
| UniProt ID | ZN816_HUMAN | ||||||||
| UniProt AC | Q0VGE8 (TFClass) | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
| ||||||||
| GTEx tissue expression atlas | ZNF816 expression | ||||||||
| ReMap ChIP-seq dataset list | ZNF816 datasets | ||||||||
| Motifs in JASPAR |
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 101.0 | 11.0 | 43.0 | 10.0 | 38.0 | 12.0 | 8.0 | 17.0 | 46.0 | 17.0 | 35.0 | 13.0 | 32.0 | 11.0 | 65.0 | 10.0 |
| 02 | 90.0 | 11.0 | 98.0 | 18.0 | 25.0 | 10.0 | 5.0 | 11.0 | 23.0 | 13.0 | 100.0 | 15.0 | 6.0 | 4.0 | 27.0 | 13.0 |
| 03 | 50.0 | 6.0 | 38.0 | 50.0 | 11.0 | 10.0 | 5.0 | 12.0 | 129.0 | 18.0 | 71.0 | 12.0 | 22.0 | 10.0 | 15.0 | 10.0 |
| 04 | 78.0 | 65.0 | 30.0 | 39.0 | 26.0 | 11.0 | 3.0 | 4.0 | 38.0 | 47.0 | 21.0 | 23.0 | 13.0 | 42.0 | 14.0 | 15.0 |
| 05 | 42.0 | 23.0 | 30.0 | 60.0 | 136.0 | 7.0 | 7.0 | 15.0 | 21.0 | 15.0 | 21.0 | 11.0 | 53.0 | 8.0 | 11.0 | 9.0 |
| 06 | 193.0 | 13.0 | 43.0 | 3.0 | 33.0 | 15.0 | 1.0 | 4.0 | 30.0 | 16.0 | 18.0 | 5.0 | 22.0 | 50.0 | 16.0 | 7.0 |
| 07 | 196.0 | 32.0 | 37.0 | 13.0 | 69.0 | 7.0 | 8.0 | 10.0 | 30.0 | 14.0 | 21.0 | 13.0 | 4.0 | 2.0 | 9.0 | 4.0 |
| 08 | 90.0 | 18.0 | 104.0 | 87.0 | 38.0 | 6.0 | 3.0 | 8.0 | 25.0 | 12.0 | 23.0 | 15.0 | 15.0 | 7.0 | 9.0 | 9.0 |
| 09 | 100.0 | 19.0 | 39.0 | 10.0 | 20.0 | 5.0 | 5.0 | 13.0 | 88.0 | 20.0 | 17.0 | 14.0 | 35.0 | 53.0 | 22.0 | 9.0 |
| 10 | 181.0 | 17.0 | 32.0 | 13.0 | 44.0 | 8.0 | 36.0 | 9.0 | 33.0 | 13.0 | 26.0 | 11.0 | 9.0 | 6.0 | 22.0 | 9.0 |
| 11 | 52.0 | 3.0 | 173.0 | 39.0 | 5.0 | 3.0 | 1.0 | 35.0 | 58.0 | 5.0 | 10.0 | 43.0 | 3.0 | 0.0 | 23.0 | 16.0 |
| 12 | 48.0 | 0.0 | 70.0 | 0.0 | 1.0 | 0.0 | 10.0 | 0.0 | 3.0 | 1.0 | 199.0 | 4.0 | 2.0 | 0.0 | 131.0 | 0.0 |
| 13 | 0.0 | 0.0 | 7.0 | 47.0 | 0.0 | 0.0 | 1.0 | 0.0 | 8.0 | 0.0 | 400.0 | 2.0 | 0.0 | 0.0 | 4.0 | 0.0 |
| 14 | 0.0 | 0.0 | 8.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 411.0 | 0.0 | 0.0 | 0.0 | 49.0 | 0.0 |
| 15 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 7.0 | 0.0 | 461.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 16 | 7.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 458.0 | 0.0 | 4.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 17 | 52.0 | 375.0 | 34.0 | 4.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 18 | 13.0 | 8.0 | 11.0 | 24.0 | 266.0 | 38.0 | 26.0 | 45.0 | 9.0 | 3.0 | 11.0 | 11.0 | 3.0 | 0.0 | 0.0 | 1.0 |
| 19 | 17.0 | 11.0 | 45.0 | 218.0 | 3.0 | 4.0 | 2.0 | 40.0 | 1.0 | 11.0 | 19.0 | 17.0 | 4.0 | 37.0 | 25.0 | 15.0 |
| 20 | 3.0 | 2.0 | 16.0 | 4.0 | 3.0 | 2.0 | 54.0 | 4.0 | 10.0 | 10.0 | 53.0 | 18.0 | 11.0 | 6.0 | 263.0 | 10.0 |
| 21 | 7.0 | 8.0 | 4.0 | 8.0 | 6.0 | 5.0 | 3.0 | 6.0 | 108.0 | 151.0 | 17.0 | 110.0 | 11.0 | 9.0 | 10.0 | 6.0 |
| 22 | 90.0 | 16.0 | 19.0 | 7.0 | 86.0 | 38.0 | 11.0 | 38.0 | 14.0 | 3.0 | 8.0 | 9.0 | 15.0 | 15.0 | 32.0 | 68.0 |
| 23 | 39.0 | 8.0 | 94.0 | 64.0 | 28.0 | 6.0 | 11.0 | 27.0 | 16.0 | 5.0 | 36.0 | 13.0 | 14.0 | 4.0 | 96.0 | 8.0 |
| 24 | 17.0 | 6.0 | 68.0 | 6.0 | 4.0 | 1.0 | 6.0 | 12.0 | 20.0 | 11.0 | 196.0 | 10.0 | 6.0 | 7.0 | 94.0 | 5.0 |
| 25 | 15.0 | 9.0 | 18.0 | 5.0 | 7.0 | 7.0 | 3.0 | 8.0 | 22.0 | 9.0 | 321.0 | 12.0 | 4.0 | 9.0 | 16.0 | 4.0 |
| 26 | 17.0 | 10.0 | 13.0 | 8.0 | 9.0 | 12.0 | 1.0 | 12.0 | 166.0 | 43.0 | 67.0 | 82.0 | 5.0 | 7.0 | 9.0 | 8.0 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 1.228 | -0.959 | 0.379 | -1.051 | 0.257 | -0.875 | -1.265 | -0.535 | 0.446 | -0.535 | 0.175 | -0.797 | 0.087 | -0.959 | 0.789 | -1.051 |
| 02 | 1.113 | -0.959 | 1.198 | -0.48 | -0.157 | -1.051 | -1.708 | -0.959 | -0.239 | -0.797 | 1.218 | -0.658 | -1.537 | -1.913 | -0.081 | -0.797 |
| 03 | 0.529 | -1.537 | 0.257 | 0.529 | -0.959 | -1.051 | -1.708 | -0.875 | 1.472 | -0.48 | 0.877 | -0.875 | -0.283 | -1.051 | -0.658 | -1.051 |
| 04 | 0.971 | 0.789 | 0.023 | 0.282 | -0.118 | -0.959 | -2.172 | -1.913 | 0.257 | 0.467 | -0.328 | -0.239 | -0.797 | 0.356 | -0.725 | -0.658 |
| 05 | 0.356 | -0.239 | 0.023 | 0.71 | 1.524 | -1.392 | -1.392 | -0.658 | -0.328 | -0.658 | -0.328 | -0.959 | 0.586 | -1.265 | -0.959 | -1.152 |
| 06 | 1.874 | -0.797 | 0.379 | -2.172 | 0.117 | -0.658 | -3.066 | -1.913 | 0.023 | -0.595 | -0.48 | -1.708 | -0.283 | 0.529 | -0.595 | -1.392 |
| 07 | 1.889 | 0.087 | 0.23 | -0.797 | 0.849 | -1.392 | -1.265 | -1.051 | 0.023 | -0.725 | -0.328 | -0.797 | -1.913 | -2.522 | -1.152 | -1.913 |
| 08 | 1.113 | -0.48 | 1.257 | 1.079 | 0.257 | -1.537 | -2.172 | -1.265 | -0.157 | -0.875 | -0.239 | -0.658 | -0.658 | -1.392 | -1.152 | -1.152 |
| 09 | 1.218 | -0.427 | 0.282 | -1.051 | -0.376 | -1.708 | -1.708 | -0.797 | 1.091 | -0.376 | -0.535 | -0.725 | 0.175 | 0.586 | -0.283 | -1.152 |
| 10 | 1.81 | -0.535 | 0.087 | -0.797 | 0.402 | -1.265 | 0.203 | -1.152 | 0.117 | -0.797 | -0.118 | -0.959 | -1.152 | -1.537 | -0.283 | -1.152 |
| 11 | 0.568 | -2.172 | 1.764 | 0.282 | -1.708 | -2.172 | -3.066 | 0.175 | 0.676 | -1.708 | -1.051 | 0.379 | -2.172 | -4.347 | -0.239 | -0.595 |
| 12 | 0.488 | -4.347 | 0.863 | -4.347 | -3.066 | -4.347 | -1.051 | -4.347 | -2.172 | -3.066 | 1.904 | -1.913 | -2.522 | -4.347 | 1.487 | -4.347 |
| 13 | -4.347 | -4.347 | -1.392 | 0.467 | -4.347 | -4.347 | -3.066 | -4.347 | -1.265 | -4.347 | 2.601 | -2.522 | -4.347 | -4.347 | -1.913 | -4.347 |
| 14 | -4.347 | -4.347 | -1.265 | -4.347 | -4.347 | -4.347 | -4.347 | -4.347 | -3.066 | -4.347 | 2.628 | -4.347 | -4.347 | -4.347 | 0.509 | -4.347 |
| 15 | -4.347 | -4.347 | -3.066 | -4.347 | -4.347 | -4.347 | -4.347 | -4.347 | -1.392 | -4.347 | 2.743 | -4.347 | -4.347 | -4.347 | -4.347 | -4.347 |
| 16 | -1.392 | -4.347 | -4.347 | -4.347 | -4.347 | -4.347 | -4.347 | -4.347 | 2.737 | -4.347 | -1.913 | -4.347 | -4.347 | -4.347 | -4.347 | -4.347 |
| 17 | 0.568 | 2.537 | 0.147 | -1.913 | -4.347 | -4.347 | -4.347 | -4.347 | -1.913 | -4.347 | -4.347 | -4.347 | -4.347 | -4.347 | -4.347 | -4.347 |
| 18 | -0.797 | -1.265 | -0.959 | -0.197 | 2.194 | 0.257 | -0.118 | 0.424 | -1.152 | -2.172 | -0.959 | -0.959 | -2.172 | -4.347 | -4.347 | -3.066 |
| 19 | -0.535 | -0.959 | 0.424 | 1.995 | -2.172 | -1.913 | -2.522 | 0.307 | -3.066 | -0.959 | -0.427 | -0.535 | -1.913 | 0.23 | -0.157 | -0.658 |
| 20 | -2.172 | -2.522 | -0.595 | -1.913 | -2.172 | -2.522 | 0.605 | -1.913 | -1.051 | -1.051 | 0.586 | -0.48 | -0.959 | -1.537 | 2.183 | -1.051 |
| 21 | -1.392 | -1.265 | -1.913 | -1.265 | -1.537 | -1.708 | -2.172 | -1.537 | 1.295 | 1.629 | -0.535 | 1.313 | -0.959 | -1.152 | -1.051 | -1.537 |
| 22 | 1.113 | -0.595 | -0.427 | -1.392 | 1.068 | 0.257 | -0.959 | 0.257 | -0.725 | -2.172 | -1.265 | -1.152 | -0.658 | -0.658 | 0.087 | 0.834 |
| 23 | 0.282 | -1.265 | 1.156 | 0.774 | -0.045 | -1.537 | -0.959 | -0.081 | -0.595 | -1.708 | 0.203 | -0.797 | -0.725 | -1.913 | 1.177 | -1.265 |
| 24 | -0.535 | -1.537 | 0.834 | -1.537 | -1.913 | -3.066 | -1.537 | -0.875 | -0.376 | -0.959 | 1.889 | -1.051 | -1.537 | -1.392 | 1.156 | -1.708 |
| 25 | -0.658 | -1.152 | -0.48 | -1.708 | -1.392 | -1.392 | -2.172 | -1.265 | -0.283 | -1.152 | 2.382 | -0.875 | -1.913 | -1.152 | -0.595 | -1.913 |
| 26 | -0.535 | -1.051 | -0.797 | -1.265 | -1.152 | -0.875 | -3.066 | -0.875 | 1.723 | 0.379 | 0.819 | 1.02 | -1.708 | -1.392 | -1.152 | -1.265 |