PWMs for transcription factors
Model |
LOGO |
Transcription factor |
Model length |
Quality |
Model rank |
Consensus |
Model release |
Data source |
Best auROC (human) |
Best auROC (mouse) |
Peak sets in benchmark (human) |
Peak sets in benchmark (mouse) |
Aligned words |
TF family |
TF subfamily |
HGNC |
MGI |
EntrezGene |
UniProt ID |
UniProt AC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PRRX1_HUMAN.H11MO.0.D | 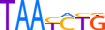 |
PRRX1 (GeneCards) |
7 | D | 0 | TAATCTG | HOCOMOCOv9 | Integrative | 4 | Paired-related HD factors {3.1.3} (TFClass) | PRRX {3.1.3.21} (TFClass) | 9142 | 5396 (SSTAR) |
PRRX1_HUMAN | P54821 (TFClass) |
|||||
| PRRX2_HUMAN.H11MO.0.C |  |
PRRX2 (GeneCards) |
7 | C | 0 | hTAATTR | HOCOMOCOv9 | Integrative | 65 | Paired-related HD factors {3.1.3} (TFClass) | PRRX {3.1.3.21} (TFClass) | 21338 | 51450 (SSTAR) |
PRRX2_HUMAN | Q99811 (TFClass) |
|||||
| PRRX1_MOUSE.H11MO.0.D | 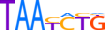 |
Prrx1 | 7 | D | 0 | TAATCTG | HOCOMOCOv9 | Integrative | 4 | Paired-related HD factors {3.1.3} (TFClass) | PRRX {3.1.3.21} (TFClass) | 97712 | 18933 (SSTAR) |
PRRX1_MOUSE | P63013 (TFClass) |
|||||
| PRRX2_MOUSE.H11MO.0.C |  |
Prrx2 | 7 | C | 0 | hTAATTR | HOCOMOCOv9 | Integrative | 65 | Paired-related HD factors {3.1.3} (TFClass) | PRRX {3.1.3.21} (TFClass) | 98218 | 20204 (SSTAR) |
PRRX2_MOUSE | Q06348 (TFClass) |