PWMs for HUMAN transcription factors from Family {1.2.6}
| Model | LOGO | Transcription factor | Model length | Quality | Model rank | Consensus | Model release | Data source | Best auROC (human) | Best auROC (mouse) | Peak sets in benchmark (human) | Peak sets in benchmark (mouse) | Aligned words | TF family | TF subfamily | HGNC | EntrezGene | UniProt ID | UniProt AC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAX_HUMAN.H11MO.0.A |  |
HUMAN:MAX | 10 | A | 0 | vnSCACGYGK | HOCOMOCOv11 | ChIP-Seq | 0.95 | 0.947 | 86 | 14 | 501 | bHLH-ZIP factors{1.2.6} | Myc / Max factors{1.2.6.5} | 6913 | 4149 | MAX_HUMAN | P61244 |
| MGAP_HUMAN.H11MO.0.D |  |
HUMAN:MGA | 16 | D | 0 | RRGTGTKAnWTWTMRC | HOCOMOCOv10 | HT-SELEX | 6178 | bHLH-ZIP factors{1.2.6}; TBX6-related factors{6.5.5} |
Mad-like factors{1.2.6.7}; MGA (MAD5){6.5.5.0.2} |
14010 | 23269 | MGAP_HUMAN | Q8IWI9 | ||||
| MITF_HUMAN.H11MO.0.A |  |
HUMAN:MITF | 10 | A | 0 | YCACGTGACh | HOCOMOCOv11 | ChIP-Seq | 0.999 | 0.824 | 28 | 3 | 516 | bHLH-ZIP factors{1.2.6} | TFE3-like factors{1.2.6.1} | 7105 | 4286 | MITF_HUMAN | O75030 |
| MLXPL_HUMAN.H11MO.0.D |  |
HUMAN:MLXIPL | 19 | D | 0 | SCAYGKGRMMMYnbSGTGb | HOCOMOCOv9 | Integrative | 11 | bHLH-ZIP factors{1.2.6} | Mondo-like factors{1.2.6.6} | 12744 | 51085 | MLXPL_HUMAN | Q9NP71 | ||||
| MLX_HUMAN.H11MO.0.D | 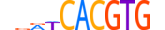 |
HUMAN:MLX | 11 | D | 0 | hdRWCACGTGn | HOCOMOCOv10 | HT-SELEX | 18958 | bHLH-ZIP factors{1.2.6} | Mondo-like factors{1.2.6.6} | 11645 | 6945 | MLX_HUMAN | Q9UH92 | ||||
| MXI1_HUMAN.H11MO.0.A | 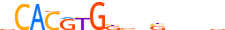 |
HUMAN:MXI1 | 15 | A | 0 | vCACRYGKvnSbbbv | HOCOMOCOv10 | ChIP-Seq | 0.878 | 0.973 | 10 | 5 | 500 | bHLH-ZIP factors{1.2.6} | Mad-like factors{1.2.6.7} | 7534 | 4601 | MXI1_HUMAN | P50539 |
| MXI1_HUMAN.H11MO.1.A |  |
HUMAN:MXI1 | 11 | A | 1 | SMCACRTGbhb | HOCOMOCOv11 | ChIP-Seq | 0.871 | 0.96 | 10 | 5 | 500 | bHLH-ZIP factors{1.2.6} | Mad-like factors{1.2.6.7} | 7534 | 4601 | MXI1_HUMAN | P50539 |
| MYCN_HUMAN.H11MO.0.A |  |
HUMAN:MYCN | 12 | A | 0 | vRvCACGTGbdv | HOCOMOCOv11 | ChIP-Seq | 0.919 | 0.899 | 32 | 10 | 501 | bHLH-ZIP factors{1.2.6} | Myc / Max factors{1.2.6.5} | 7559 | 4613 | MYCN_HUMAN | P04198 |
| MYC_HUMAN.H11MO.0.A |  |
HUMAN:MYC | 11 | A | 0 | vMCACGTGShb | HOCOMOCOv11 | ChIP-Seq | 0.947 | 0.948 | 265 | 126 | 501 | bHLH-ZIP factors{1.2.6} | Myc / Max factors{1.2.6.5} | 7553 | 4609 | MYC_HUMAN | P01106 |
| SRBP1_HUMAN.H11MO.0.A |  |
HUMAN:SREBF1 | 11 | A | 0 | dRKSRSGTGAb | HOCOMOCOv11 | ChIP-Seq | 0.92 | 0.67 | 5 | 23 | 500 | bHLH-ZIP factors{1.2.6} | SREBP factors{1.2.6.3} | 11289 | 6720 | SRBP1_HUMAN | P36956 |
| SRBP2_HUMAN.H11MO.0.B |  |
HUMAN:SREBF2 | 13 | B | 0 | vvvWGGvSWGRnb | HOCOMOCOv9 | Integrative | 2363 | bHLH-ZIP factors{1.2.6} | SREBP factors{1.2.6.3} | 11290 | 6721 | SRBP2_HUMAN | Q12772 | ||||
| TFAP4_HUMAN.H11MO.0.A |  |
HUMAN:TFAP4 | 10 | A | 0 | hCAGCTGdKb | HOCOMOCOv11 | ChIP-Seq | 0.923 | 7 | 503 | bHLH-ZIP factors{1.2.6} | AP-4{1.2.6.4} | 11745 | 7023 | TFAP4_HUMAN | Q01664 | ||
| TFE3_HUMAN.H11MO.0.B |  |
HUMAN:TFE3 | 10 | B | 0 | dGTCACGTGA | HOCOMOCOv11 | ChIP-Seq | 0.483 | 1.0 | 2 | 30 | 478 | bHLH-ZIP factors{1.2.6} | TFE3-like factors{1.2.6.1} | 11752 | 7030 | TFE3_HUMAN | P19532 |
| TFEB_HUMAN.H11MO.0.C | 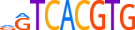 |
HUMAN:TFEB | 9 | C | 0 | dGTCACGTG | HOCOMOCOv9 | Integrative | 301 | bHLH-ZIP factors{1.2.6} | TFE3-like factors{1.2.6.1} | 11753 | 7942 | TFEB_HUMAN | P19484 | ||||
| USF1_HUMAN.H11MO.0.A |  |
HUMAN:USF1 | 12 | A | 0 | vGTCACGTGvbb | HOCOMOCOv11 | ChIP-Seq | 0.998 | 0.98 | 59 | 7 | 485 | bHLH-ZIP factors{1.2.6} | USF factors{1.2.6.2} | 12593 | 7391 | USF1_HUMAN | P22415 |
| USF2_HUMAN.H11MO.0.A |  |
HUMAN:USF2 | 19 | A | 0 | vvGTCACGTGvbvvbbvbb | HOCOMOCOv10 | ChIP-Seq | 0.998 | 0.998 | 23 | 10 | 501 | bHLH-ZIP factors{1.2.6} | USF factors{1.2.6.2} | 12594 | 7392 | USF2_HUMAN | Q15853 |